Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *Unverified author* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_hierarchicalclustering.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
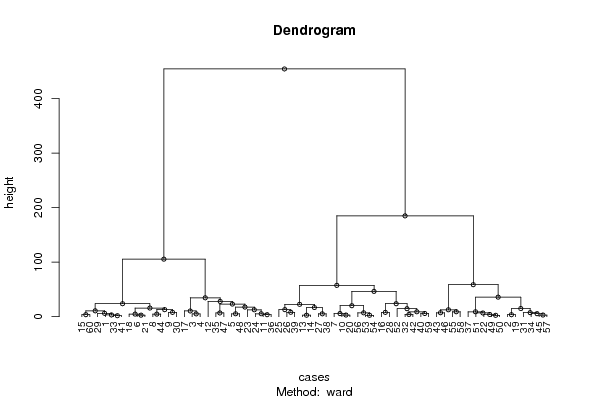
| Title produced by software | Hierarchical Clustering | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 05 Nov 2007 03:02:49 -0700 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2007/Nov/05/c9atmdy2xi5ldel1194256844.htm/, Retrieved Mon, 29 Apr 2024 06:55:20 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=454, Retrieved Mon, 29 Apr 2024 06:55:20 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | WS4 Q2 dendrogram | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 289 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Bivariate Kernel Density Estimation] [Q1 Bivariate Dens...] [2007-11-03 14:50:57] [e2ec4dc832988c648c062d4cdc574d44] - RMPD [Hierarchical Clustering] [WS4 Q2 dendrogram] [2007-11-05 10:02:49] [d41d8cd98f00b204e9800998ecf8427e] [Current] F R D [Hierarchical Clustering] [eda various topic...] [2008-11-04 19:43:54] [077ffec662d24c06be4c491541a44245] F D [Hierarchical Clustering] [] [2008-11-09 18:17:28] [4c8dfb519edec2da3492d7e6be9a5685] F RMPD [Star Plot] [] [2008-11-09 18:25:13] [4c8dfb519edec2da3492d7e6be9a5685] F P [Hierarchical Clustering] [Hierarchical clus...] [2008-11-11 16:01:43] [73d6180dc45497329efd1b6934a84aba] F P [Hierarchical Clustering] [Hierarchical Clus...] [2008-11-11 17:33:24] [6816386b1f3c2f6c0c9f2aa1e5bc9362] F [Hierarchical Clustering] [] [2008-11-11 19:45:33] [a7a7b7de998247cdf0f65ef79d563d66] - [Hierarchical Clustering] [] [2008-11-21 17:22:54] [888addc516c3b812dd7be4bd54caa358] - [Hierarchical Clustering] [Various EDA Topic...] [2008-11-22 09:58:44] [33f4701c7363e8b81858dafbf0350eed] F RMPD [Star Plot] [various EDA topic...] [2008-11-04 19:47:59] [077ffec662d24c06be4c491541a44245] F P [Star Plot] [Star plot] [2008-11-11 16:04:19] [73d6180dc45497329efd1b6934a84aba] - P [Star Plot] [Starplot] [2008-11-11 17:35:54] [6816386b1f3c2f6c0c9f2aa1e5bc9362] - RMPD [Pearson Correlation] [various eda topics] [2008-11-04 20:17:00] [077ffec662d24c06be4c491541a44245] - RMPD [Pearson Correlation] [various eda topic...] [2008-11-04 20:17:00] [077ffec662d24c06be4c491541a44245] - RMPD [Pearson Correlation] [various eda topic...] [2008-11-04 20:17:00] [077ffec662d24c06be4c491541a44245] F RMPD [Box-Cox Linearity Plot] [] [2008-11-04 20:21:52] [077ffec662d24c06be4c491541a44245] F PD [Box-Cox Linearity Plot] [] [2008-11-09 18:32:27] [4c8dfb519edec2da3492d7e6be9a5685] F P [Box-Cox Linearity Plot] [Box-cox linearity...] [2008-11-11 16:06:09] [73d6180dc45497329efd1b6934a84aba] F P [Box-Cox Linearity Plot] [Box-Cox linearity...] [2008-11-11 17:37:44] [6816386b1f3c2f6c0c9f2aa1e5bc9362] F [Box-Cox Linearity Plot] [] [2008-11-11 19:54:20] [a7a7b7de998247cdf0f65ef79d563d66] - [Box-Cox Linearity Plot] [] [2008-11-21 17:56:22] [888addc516c3b812dd7be4bd54caa358] - RMPD [Box-Cox Normality Plot] [various eda topic...] [2008-11-04 20:30:40] [077ffec662d24c06be4c491541a44245] F D [Box-Cox Normality Plot] [various eda topic...] [2008-11-04 20:48:12] [077ffec662d24c06be4c491541a44245] F PD [Box-Cox Normality Plot] [] [2008-11-09 18:41:47] [4c8dfb519edec2da3492d7e6be9a5685] F P [Box-Cox Normality Plot] [Box-Cox Normality...] [2008-11-11 16:08:06] [73d6180dc45497329efd1b6934a84aba] F [Box-Cox Normality Plot] [Box-Cox Normality...] [2008-11-11 17:40:11] [6816386b1f3c2f6c0c9f2aa1e5bc9362] F [Box-Cox Normality Plot] [] [2008-11-11 20:04:58] [a7a7b7de998247cdf0f65ef79d563d66] - [Box-Cox Normality Plot] [] [2008-11-21 18:02:08] [888addc516c3b812dd7be4bd54caa358] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
99,0 101,5 108,4 115,4 126,6 117,0 106,9 93,9 103,8 107,1 89,8 100,8 99,3 93,4 110,6 99,2 101,5 104,0 108,3 110,4 112,6 105,6 105,9 107,3 99,5 108,4 98,9 107,4 113,9 109,8 93,1 86,1 104,9 88,1 69,4 102,2 110,7 101,2 123,9 113,1 100,5 124,9 99,6 98,0 112,7 93,6 106,6 121,9 98,6 90,1 100,6 99,6 96,9 104,3 114,3 125,9 120,4 107,8 112,0 107,5 101,2 100,0 102,9 112,5 123,9 125,6 100,5 79,8 107,5 93,9 83,4 108,8 116,2 113,6 128,4 112,0 112,9 121,1 106,4 104,0 119,5 95,7 109,9 128,7 96,0 99,0 108,7 95,8 106,3 105,5 103,0 128,9 119,8 102,2 111,1 111,3 98,4 102,9 110,6 111,4 130,0 120,1 86,6 87,0 97,5 91,3 87,5 107,7 107,9 117,6 127,3 101,8 103,4 117,2 104,4 110,8 119,8 93,4 112,6 116,2 100,1 102,5 111,0 98,5 112,4 112,4 112,9 135,6 130,6 101,4 105,1 109,1 107,1 127,7 118,8 110,8 137,0 123,9 90,3 91,0 101,6 95,5 90,5 112,8 111,4 122,4 128,0 113,0 123,3 129,6 107,5 124,3 125,8 95,9 120,0 119,5 106,3 118,1 115,7 105,2 119,0 113,6 117,2 142,7 129,7 106,9 123,6 112,0 108,2 129,6 116,8 110,0 146,9 126,3 96,1 108,7 112,9 100,6 99,4 115,9 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = ward ; par2 = ALL ; par3 = FALSE ; par4 = FALSE ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = ward ; par2 = ALL ; par3 = FALSE ; par4 = FALSE ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par3 <- as.logical(par3) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||