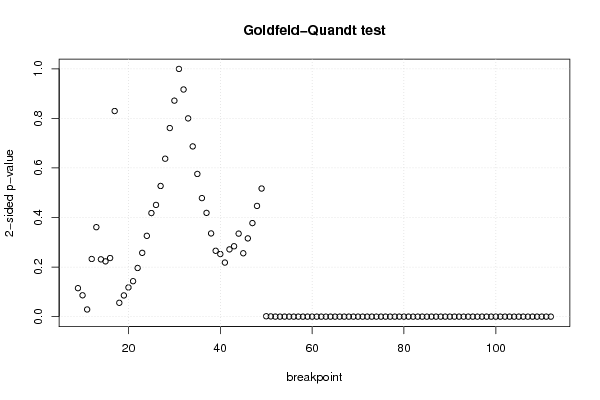
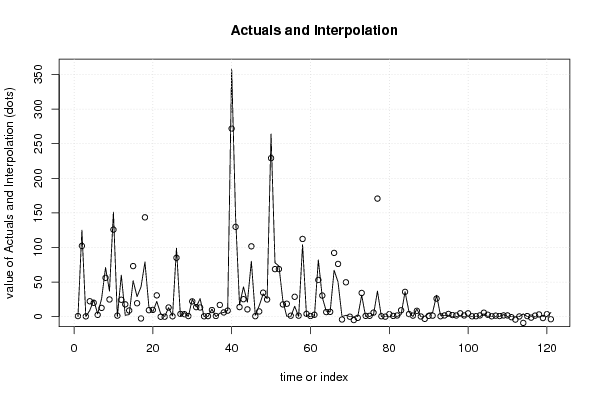
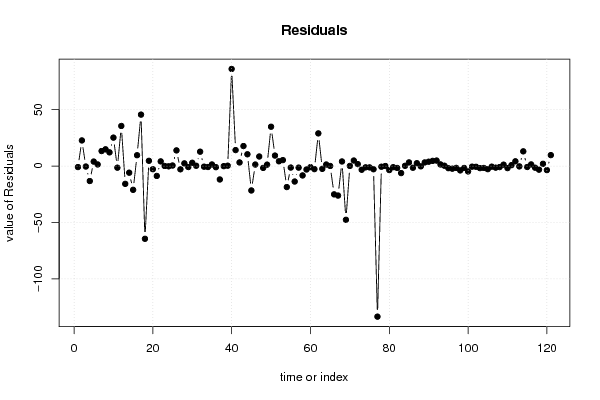
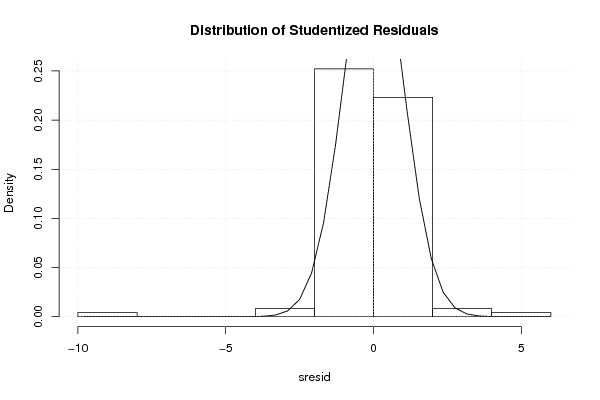
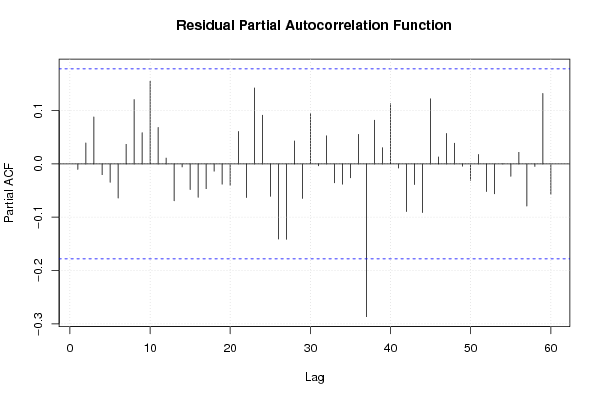
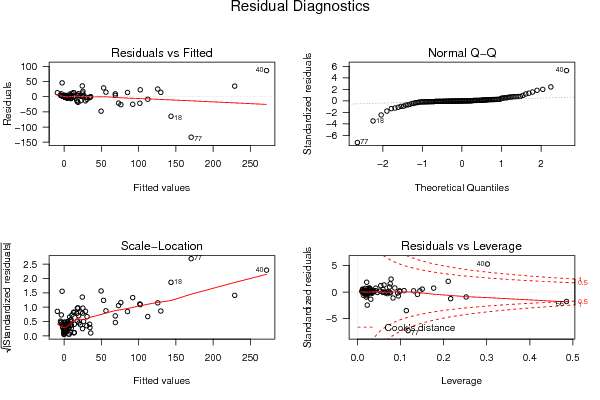
0,00 0,00 0,00 0,00 180,00 21,00
125,00 121,00 11,00 0,00 326,00 21,00
0,00 0,00 1,00 0,00 332,00 21,00
9,00 28,00 0,00 0,00 274,00 21,00
24,00 26,00 1,00 0,00 459,00 21,00
4,00 1,00 3,00 2,00 263,00 21,00
26,00 12,00 5,00 2,00 243,00 21,00
71,00 66,00 11,00 12,00 365,00 21,00
37,00 30,00 7,00 16,00 300,00 21,00
151,00 155,00 8,00 9,00 262,00 21,00
0,00 0,00 0,00 0,00 100,00 21,00
60,00 27,00 3,00 0,00 132,00 21,00
2,00 2,00 25,00 2,00 206,00 21,00
3,00 0,00 12,00 0,00 130,00 21,00
52,00 99,00 4,00 17,00 864,00 21,00
29,00 25,00 2,00 0,00 526,00 21,00
43,00 0,00 2,00 0,00 852,00 21,00
79,00 180,00 10,00 5,00 827,00 21,00
14,00 13,00 0,00 0,00 431,00 21,00
7,00 12,00 2,00 3,00 350,00 21,00
22,00 40,00 0,00 4,00 294,00 21,00
4,00 2,00 7,00 34,00 187,00 21,00
0,00 0,00 0,00 0,00 314,00 21,00
13,00 15,00 2,00 7,00 82,00 21,00
1,00 0,00 3,00 2,00 440,00 21,00
99,00 105,00 6,00 0,00 516,00 21,00
1,00 3,00 0,00 0,00 90,00 21,00
6,00 2,00 2,00 0,00 194,00 21,00
0,00 0,00 0,00 0,00 183,00 20,00
25,00 24,00 11,00 12,00 500,00 20,00
14,00 17,00 2,00 6,00 263,00 20,00
26,00 12,00 8,00 8,00 261,00 20,00
0,00 0,00 0,00 0,00 228,00 20,00
0,00 0,00 0,00 0,00 187,00 20,00
11,00 13,00 0,00 3,00 310,00 20,00
0,00 0,00 0,00 0,00 167,00 20,00
5,00 22,00 1,00 6,00 282,00 20,00
6,00 8,00 0,00 1,00 305,00 20,00
9,00 4,00 13,00 7,00 490,00 20,00
358,00 342,00 14,00 28,00 448,00 20,00
144,00 164,00 8,00 7,00 747,00 20,00
17,00 19,00 0,00 1,00 441,00 20,00
43,00 39,00 1,00 27,00 387,00 20,00
21,00 12,00 4,00 5,00 366,00 20,00
80,00 125,00 24,00 47,00 660,00 20,00
2,00 0,00 4,00 2,00 510,00 20,00
16,00 8,00 2,00 2,00 255,00 20,00
33,00 41,00 7,00 11,00 286,00 20,00
26,00 33,00 0,00 2,00 425,00 20,00
264,00 279,00 19,00 15,00 296,00 20,00
78,00 77,00 15,00 2,00 470,00 20,00
73,00 84,00 4,00 6,00 134,00 20,00
23,00 22,00 0,00 2,00 195,00 20,00
0,00 22,00 0,00 0,00 123,00 20,00
0,00 0,00 0,00 0,00 115,00 20,00
15,00 30,00 17,00 21,00 507,00 20,00
0,00 0,00 0,00 0,00 109,00 20,00
104,00 129,00 24,00 12,00 618,00 20,00
1,00 0,00 6,00 0,00 274,00 20,00
0,00 0,00 0,00 0,00 168,00 20,00
0,00 0,00 3,00 0,00 190,00 20,00
82,00 54,00 15,00 2,00 126,00 20,00
28,00 38,00 1,00 1,00 294,00 20,00
8,00 9,00 1,00 2,00 399,00 20,00
7,00 7,00 3,00 2,00 317,00 20,00
67,00 131,00 7,00 75,00 331,00 20,00
50,00 95,00 3,00 3,00 308,00 20,00
0,00 0,00 3,00 0,00 1144,00 20,00
2,00 63,00 2,00 1,00 452,00 20,00
0,00 0,00 0,00 0,00 322,00 20,00
0,00 0,00 0,00 0,00 990,00 20,00
0,00 0,00 0,00 0,00 547,00 20,00
31,00 43,00 3,00 2,00 464,00 20,00
0,00 0,00 0,00 0,00 143,00 20,00
0,00 0,00 0,00 0,00 138,00 20,00
3,00 0,00 12,00 0,00 560,00 20,00
37,00 211,00 10,00 7,00 400,00 20,00
0,00 0,00 0,00 0,00 221,00 20,00
0,00 0,00 0,00 0,00 310,00 20,00
0,00 0,00 3,00 0,00 74,00 20,00
0,00 0,00 0,00 0,00 162,00 20,00
0,00 0,00 0,00 0,00 67,00 20,00
3,00 11,00 0,00 1,00 198,00 20,00
36,00 46,00 1,00 7,00 259,00 20,00
7,00 3,00 1,00 2,00 140,00 20,00
0,00 0,00 0,00 0,00 94,00 20,00
11,00 10,00 1,00 1,00 285,00 19,00
0,00 0,00 0,00 0,00 270,00 19,00
0,00 0,00 0,00 0,00 755,00 19,00
5,00 0,00 1,00 3,00 148,00 19,00
6,00 2,00 0,00 3,00 244,00 19,00
31,00 31,00 7,00 0,00 697,00 18,00
2,00 0,00 0,00 0,00 226,00 18,00
2,00 3,00 0,00 0,00 400,00 18,00
2,00 2,00 2,00 2,00 105,00 17,00
0,00 0,00 6,00 5,00 376,00 17,00
0,00 0,00 0,00 0,00 79,00 17,00
1,00 4,00 1,00 0,00 160,00 17,00
0,00 0,00 0,00 0,00 70,00 17,00
0,00 0,00 5,00 0,00 87,00 17,00
0,00 0,00 0,00 0,00 234,00 17,00
0,00 0,00 0,00 0,00 223,00 16,00
0,00 0,00 7,00 0,00 682,00 16,00
4,00 6,00 0,00 1,00 149,00 16,00
0,00 0,00 3,00 0,00 195,00 16,00
0,00 0,00 0,00 0,00 236,00 16,00
0,00 0,00 0,00 0,00 112,00 16,00
0,00 0,00 0,00 0,00 184,00 16,00
3,00 0,00 0,00 0,00 65,00 16,00
0,00 0,00 0,00 0,00 62,00 15,00
0,00 0,00 0,00 0,00 424,00 15,00
0,00 0,00 0,00 0,00 906,00 15,00
0,00 0,00 0,00 0,00 271,00 15,00
4,00 1,00 0,00 34,00 712,00 14,00
0,00 0,00 0,00 0,00 200,00 13,00
0,00 0,00 0,00 0,00 520,00 13,00
0,00 0,00 0,00 0,00 113,00 13,00
0,00 2,00 0,00 0,00 89,00 13,00
0,00 0,00 0,00 0,00 600,00 12,00
0,00 3,00 0,00 0,00 148,00 12,00
6,00 0,00 5,00 13,00 912,00 12,00
|