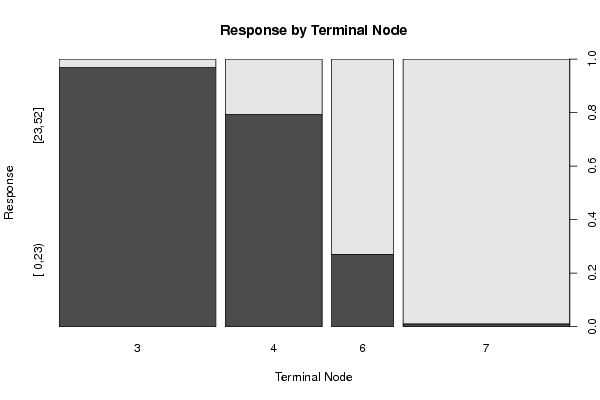Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_regression_trees1.wasp | ||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Recursive Partitioning (Regression Trees) | ||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 19 Dec 2011 14:16:47 -0500 | ||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2011/Dec/19/t1324324491au9c9cluw7d90n3.htm/, Retrieved Wed, 15 May 2024 07:51:26 +0000 | ||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=157652, Retrieved Wed, 15 May 2024 07:51:26 +0000 | |||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 105 | ||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||
| - [Recursive Partitioning (Regression Trees)] [] [2010-12-05 18:59:57] [b98453cac15ba1066b407e146608df68] - PD [Recursive Partitioning (Regression Trees)] [WS 10 - recursive...] [2010-12-11 16:07:41] [033eb2749a430605d9b2be7c4aac4a0c] - [Recursive Partitioning (Regression Trees)] [] [2010-12-13 18:24:09] [d7b28a0391ab3b2ddc9f9fba95a43f33] - [Recursive Partitioning (Regression Trees)] [] [2010-12-25 21:51:47] [2e1e44f0ae3cb9513dc28781dfdb387b] - PD [Recursive Partitioning (Regression Trees)] [Recursive Partiti...] [2011-12-15 19:16:06] [9bc4436833541bde47df2e8d3b08804c] - PD [Recursive Partitioning (Regression Trees)] [with categorizati...] [2011-12-19 19:16:47] [f914a0f804421ae312123c83c378984e] [Current] | |||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||
30 112285 24188 146283 144 145 28 84786 18273 98364 103 101 38 83123 14130 86146 98 98 30 101193 32287 96933 135 132 22 38361 8654 79234 61 60 26 68504 9245 42551 39 38 25 119182 33251 195663 150 144 18 22807 1271 6853 5 5 11 17140 5279 21529 28 28 26 116174 27101 95757 84 84 25 57635 16373 85584 80 79 38 66198 19716 143983 130 127 44 71701 17753 75851 82 78 30 57793 9028 59238 60 60 40 80444 18653 93163 131 131 34 53855 8828 96037 84 84 47 97668 29498 151511 140 133 30 133824 27563 136368 151 150 31 101481 18293 112642 91 91 23 99645 22530 94728 138 132 36 114789 15977 105499 150 136 36 99052 35082 121527 124 124 30 67654 16116 127766 119 118 25 65553 15849 98958 73 70 39 97500 16026 77900 110 107 34 69112 26569 85646 123 119 31 82753 24785 98579 90 89 31 85323 17569 130767 116 112 33 72654 23825 131741 113 108 25 30727 7869 53907 56 52 33 77873 14975 178812 115 112 35 117478 37791 146761 119 116 42 74007 9605 82036 129 123 43 90183 27295 163253 127 125 30 61542 2746 27032 27 27 33 101494 34461 171975 175 162 13 27570 8098 65990 35 32 32 55813 4787 86572 64 64 36 79215 24919 159676 96 92 0 1423 603 1929 0 0 28 55461 16329 85371 84 83 14 31081 12558 58391 41 41 17 22996 7784 31580 47 47 32 83122 28522 136815 126 120 30 70106 22265 120642 105 105 35 60578 14459 69107 80 79 20 39992 14526 50495 70 65 28 79892 22240 108016 73 70 28 49810 11802 46341 57 55 39 71570 7623 78348 40 39 34 100708 11912 79336 68 67 26 33032 7935 56968 21 21 39 82875 18220 93176 127 127 39 139077 19199 161632 154 152 33 71595 19918 87850 116 113 28 72260 21884 127969 102 99 4 5950 2694 15049 7 7 39 115762 15808 155135 148 141 18 32551 3597 25109 21 21 14 31701 5296 45824 35 35 29 80670 25239 102996 112 109 44 143558 29801 160604 137 133 21 117105 18450 158051 135 123 16 23789 7132 44547 26 26 28 120733 34861 162647 230 230 35 105195 35940 174141 181 166 28 73107 16688 60622 71 68 38 132068 24683 179566 147 147 23 149193 46230 184301 190 179 36 46821 10387 75661 64 61 32 87011 21436 96144 105 101 29 95260 30546 129847 107 108 25 55183 19746 117286 94 90 27 106671 15977 71180 116 114 36 73511 22583 109377 106 103 28 92945 17274 85298 143 142 23 78664 16469 73631 81 79 40 70054 14251 86767 89 88 23 22618 3007 23824 26 25 40 74011 16851 93487 84 83 28 83737 21113 82981 113 113 34 69094 17401 73815 120 118 33 93133 23958 94552 110 110 28 95536 23567 132190 134 129 34 225920 13065 128754 54 51 30 62133 15358 66363 96 93 33 61370 14587 67808 78 76 22 43836 12770 61724 51 49 38 106117 24021 131722 121 118 26 38692 9648 68580 38 38 35 84651 20537 106175 145 141 8 56622 7905 55792 59 58 24 15986 4527 25157 27 27 29 95364 30495 76669 91 91 20 26706 7117 57283 48 48 29 89691 17719 105805 68 63 45 67267 27056 129484 58 56 37 126846 33473 72413 150 144 33 41140 9758 87831 74 73 33 102860 21115 96971 181 168 25 51715 7236 71299 65 64 32 55801 13790 77494 97 97 29 111813 32902 120336 121 117 28 120293 25131 93913 99 100 28 138599 30910 136048 152 149 31 161647 35947 181248 188 187 52 115929 29848 146123 138 127 21 24266 6943 32036 40 37 24 162901 42705 186646 254 245 41 109825 31808 102255 87 87 33 129838 26675 168237 178 177 32 37510 8435 64219 51 49 19 43750 7409 19630 49 49 20 40652 14993 76825 73 73 31 87771 36867 115338 176 177 31 85872 33835 109427 94 94 32 89275 24164 118168 120 117 18 44418 12607 84845 66 60 23 192565 22609 153197 56 55 17 35232 5892 29877 39 39 20 40909 17014 63506 66 64 12 13294 5394 22445 27 26 17 32387 9178 47695 65 64 30 140867 6440 68370 58 58 31 120662 21916 146304 98 95 10 21233 4011 38233 25 25 13 44332 5818 42071 26 26 22 61056 18647 50517 77 76 42 101338 20556 103950 130 129 1 1168 238 5841 11 11 9 13497 70 2341 2 2 32 65567 22392 84396 101 101 11 25162 3913 24610 31 28 25 32334 12237 35753 36 36 36 40735 8388 55515 120 89 31 91413 22120 209056 195 193 0 855 338 6622 4 4 24 97068 11727 115814 89 84 13 44339 3704 11609 24 23 8 14116 3988 13155 39 39 13 10288 3030 18274 14 14 19 65622 13520 72875 78 78 18 16563 1421 10112 15 14 33 76643 20923 142775 106 101 40 110681 20237 68847 83 82 22 29011 3219 17659 24 24 38 92696 3769 20112 37 36 24 94785 12252 61023 77 75 8 8773 1888 13983 16 16 35 83209 14497 65176 56 55 43 93815 28864 132432 132 131 43 86687 21721 112494 144 131 14 34553 4821 45109 40 39 41 105547 33644 170875 153 144 38 103487 15923 180759 143 139 45 213688 42935 214921 220 211 31 71220 18864 100226 79 78 13 23517 4977 32043 50 50 28 56926 7785 54454 39 39 31 91721 17939 78876 95 90 40 115168 23436 170745 169 166 30 111194 325 6940 12 12 16 51009 13539 49025 63 57 37 135777 34538 122037 134 133 30 51513 12198 53782 69 69 35 74163 26924 127748 119 119 32 51633 12716 86839 119 119 27 75345 8172 44830 75 65 20 33416 10855 77395 63 61 18 83305 11932 89324 55 49 31 98952 14300 103300 103 101 31 102372 25515 112283 197 196 21 37238 2805 10901 16 15 39 103772 29402 120691 140 136 41 123969 16440 58106 89 89 13 27142 11221 57140 40 40 32 135400 28732 122422 125 123 18 21399 5250 25899 21 21 39 130115 28608 139296 167 163 14 24874 8092 52678 32 29 7 34988 4473 23853 36 35 17 45549 1572 17306 13 13 0 6023 2065 7953 5 5 30 64466 14817 89455 96 96 37 54990 16714 147866 151 151 0 1644 556 4245 6 6 5 6179 2089 21509 13 13 1 3926 2658 7670 3 3 16 32755 10695 66675 57 56 32 34777 1669 14336 23 23 24 73224 16267 53608 61 57 17 27114 7768 30059 21 14 11 20760 7252 29668 43 43 24 37636 6387 22097 20 20 22 65461 18715 96841 82 72 12 30080 7936 41907 90 87 19 24094 8643 27080 25 21 13 69008 7294 35885 60 56 17 54968 4570 41247 61 59 15 46090 7185 28313 85 82 16 27507 10058 36845 43 43 24 10672 2342 16548 25 25 15 34029 8509 36134 41 38 17 46300 13275 55764 26 25 18 24760 6816 28910 38 38 20 18779 1930 13339 12 12 16 21280 8086 25319 29 29 16 40662 10737 66956 49 47 18 28987 8033 47487 46 45 22 22827 7058 52785 41 40 8 18513 6782 44683 31 30 17 30594 5401 35619 41 41 18 24006 6521 21920 26 25 16 27913 10856 45608 23 23 23 42744 2154 7721 14 14 22 12934 6117 20634 16 16 13 22574 5238 29788 25 26 13 41385 4820 31931 21 21 16 18653 5615 37754 32 27 16 18472 4272 32505 9 9 20 30976 8702 40557 35 33 22 63339 15340 94238 42 42 17 25568 8030 44197 68 68 18 33747 9526 43228 32 32 17 4154 1278 4103 6 6 12 19474 4236 44144 68 67 7 35130 3023 32868 33 33 17 39067 7196 27640 84 77 14 13310 3394 14063 46 46 23 65892 6371 28990 30 30 17 4143 1574 4694 0 0 14 28579 9620 42648 36 36 15 51776 6978 64329 47 46 17 21152 4911 21928 20 18 21 38084 8645 25836 50 48 18 27717 8987 22779 30 29 18 32928 5544 40820 30 28 17 11342 3083 27530 34 34 17 19499 6909 32378 33 33 16 16380 3189 10824 34 34 15 36874 6745 39613 37 33 21 48259 16724 60865 83 80 16 16734 4850 19787 32 32 14 28207 7025 20107 30 30 15 30143 6047 36605 43 41 17 41369 7377 40961 41 41 15 45833 9078 48231 51 51 15 29156 4605 39725 19 18 10 35944 3238 21455 37 34 6 36278 8100 23430 33 31 22 45588 9653 62991 41 39 21 45097 8914 49363 54 54 1 3895 786 9604 14 14 18 28394 6700 24552 25 24 17 18632 5788 31493 25 24 4 2325 593 3439 8 8 10 25139 4506 19555 26 26 16 27975 6382 21228 20 19 16 14483 5621 23177 11 11 9 13127 3997 22094 14 14 16 5839 520 2342 3 1 17 24069 8891 38798 40 39 7 3738 999 3255 5 5 15 18625 7067 24261 38 37 14 36341 4639 18511 32 32 14 24548 5654 40798 41 38 18 21792 6928 28893 46 47 12 26263 1514 21425 47 47 16 23686 9238 50276 37 37 21 49303 8204 37643 51 51 19 25659 5926 30377 49 45 16 28904 5785 27126 21 21 1 2781 4 13 1 1 16 29236 5930 42097 44 42 10 19546 3710 24451 26 26 19 22818 705 14335 21 21 12 32689 443 5084 4 4 2 5752 2416 9927 10 10 14 22197 7747 43527 43 43 17 20055 5432 27184 34 34 19 25272 4913 21610 32 31 14 82206 2650 20484 20 19 11 32073 2370 20156 34 34 4 5444 775 6012 6 6 16 20154 5576 18475 12 11 20 36944 1352 12645 24 24 12 8019 3080 11017 16 16 15 30884 10205 37623 72 72 16 19540 6095 35873 27 21 | |||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||
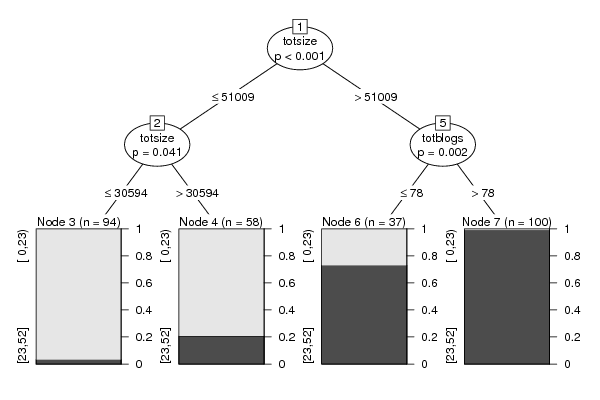
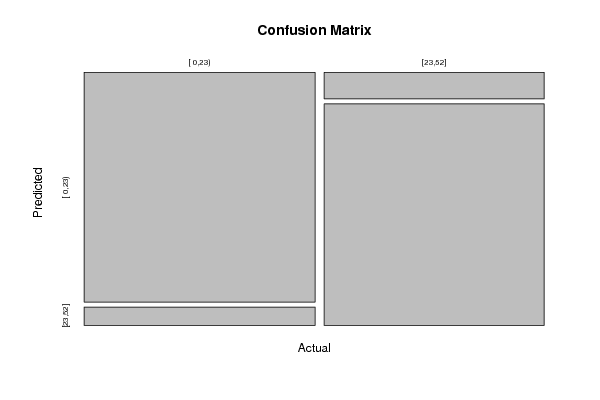
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = quantiles ; par3 = 2 ; par4 = no ; | |||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = quantiles ; par3 = 2 ; par4 = no ; | |||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||
library(party) | |||||||||||||||||||||||||||||||||||||||||||||