| Multiple Linear Regression - Estimated Regression Equation |
| Carbon_Footprint[t] = -1.26537 + 0.000122018`Population_(millions)`[t] + 3.20593HDI[t] + 6.38132e-05GDP_per_Capita[t] + 0.00416326Total_Biocapacity[t] + e[t] |
| Multiple Linear Regression - Ordinary Least Squares | |||||
| Variable | Parameter | S.D. | T-STAT H0: parameter = 0 | 2-tail p-value | 1-tail p-value |
| (Intercept) | -1.265 | 0.4496 | -2.8140e+00 | 0.005512 | 0.002756 |
| `Population_(millions)` | +0.000122 | 0.0005486 | +2.2240e-01 | 0.8243 | 0.4121 |
| HDI | +3.206 | 0.7185 | +4.4620e+00 | 1.542e-05 | 7.711e-06 |
| GDP_per_Capita | +6.381e-05 | 5.702e-06 | +1.1190e+01 | 9.532e-22 | 4.766e-22 |
| Total_Biocapacity | +0.004163 | 0.009001 | +4.6250e-01 | 0.6444 | 0.3222 |
| Multiple Linear Regression - Regression Statistics | |
| Multiple R | 0.8461 |
| R-squared | 0.7159 |
| Adjusted R-squared | 0.7086 |
| F-TEST (value) | 98.89 |
| F-TEST (DF numerator) | 4 |
| F-TEST (DF denominator) | 157 |
| p-value | 0 |
| Multiple Linear Regression - Residual Statistics | |
| Residual Standard Deviation | 1.047 |
| Sum Squared Residuals | 172.1 |
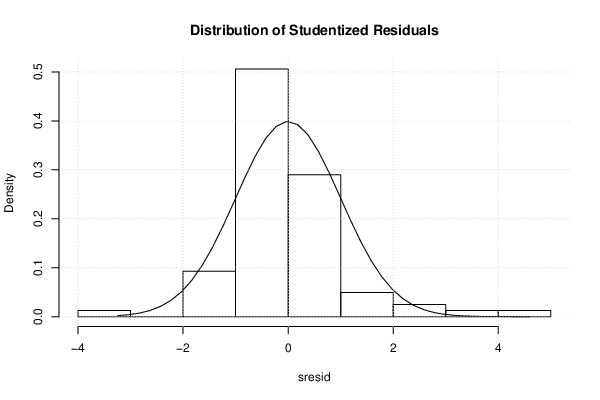
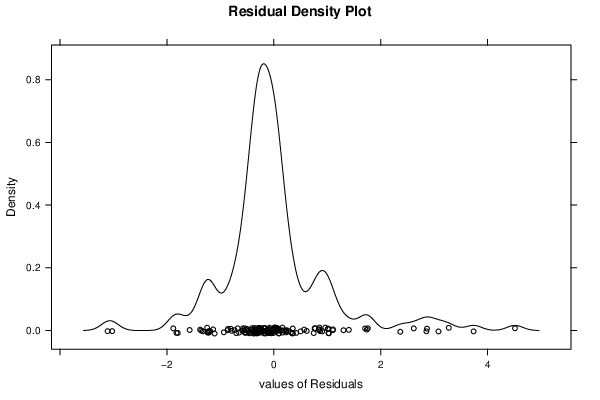
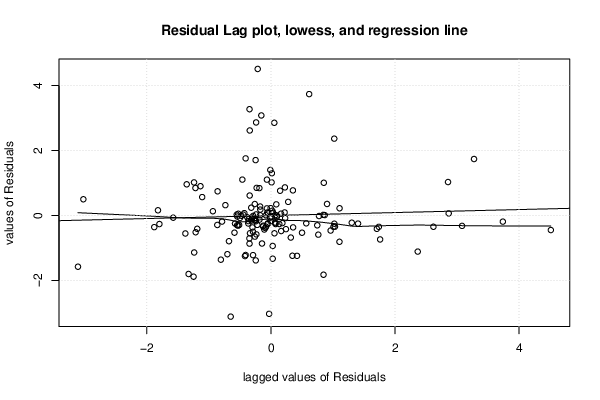
| Menu of Residual Diagnostics | |
| Description | Link |
| Histogram | Compute |
| Central Tendency | Compute |
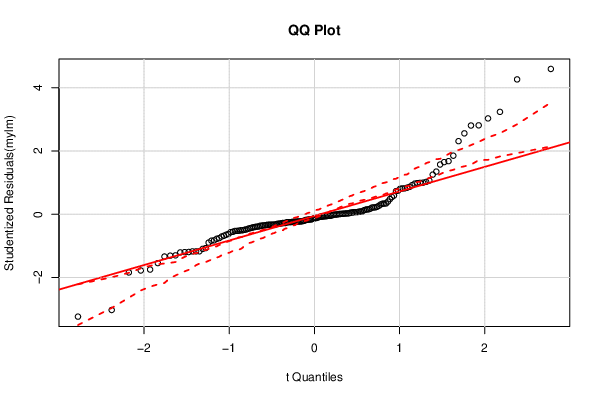
| QQ Plot | Compute |
| Kernel Density Plot | Compute |
| Skewness/Kurtosis Test | Compute |
| Skewness-Kurtosis Plot | Compute |
| Harrell-Davis Plot | Compute |
| Bootstrap Plot -- Central Tendency | Compute |
| Blocked Bootstrap Plot -- Central Tendency | Compute |
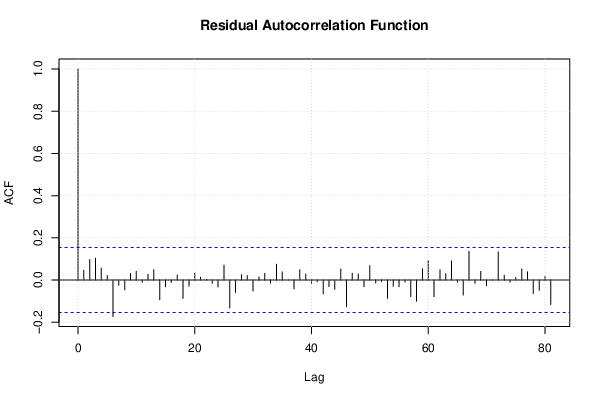
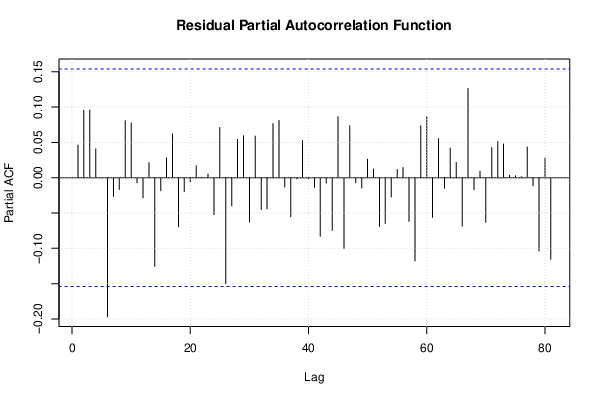
| (Partial) Autocorrelation Plot | Compute |
| Spectral Analysis | Compute |
| Tukey lambda PPCC Plot | Compute |
| Box-Cox Normality Plot | Compute |
| Summary Statistics | Compute |
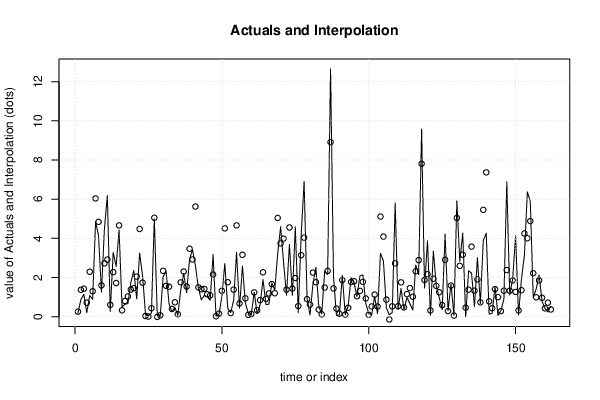
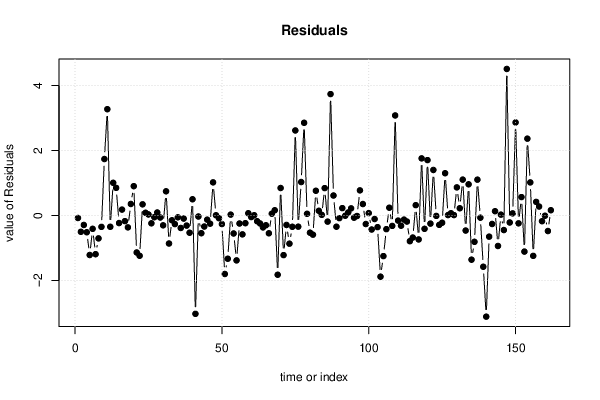
| Multiple Linear Regression - Actuals, Interpolation, and Residuals | |||
| Time or Index | Actuals | Interpolation Forecast | Residuals Prediction Error |
| 1 | 0.18 | 0.2543 | -0.0743 |
| 2 | 0.87 | 1.37 | -0.4996 |
| 3 | 1.14 | 1.429 | -0.2887 |
| 4 | 0.2 | 0.7126 | -0.5126 |
| 5 | 1.08 | 2.293 | -1.213 |
| 6 | 0.89 | 1.298 | -0.4077 |
| 7 | 4.85 | 6.038 | -1.188 |
| 8 | 4.14 | 4.842 | -0.7016 |
| 9 | 1.25 | 1.597 | -0.3472 |
| 10 | 4.46 | 2.72 | 1.74 |
| 11 | 6.19 | 2.917 | 3.273 |
| 12 | 0.26 | 0.6051 | -0.3451 |
| 13 | 3.28 | 2.271 | 1.009 |
| 14 | 2.57 | 1.718 | 0.8523 |
| 15 | 4.43 | 4.661 | -0.2306 |
| 16 | 0.51 | 0.326 | 0.184 |
| 17 | 0.63 | 0.801 | -0.171 |
| 18 | 0.67 | 1.033 | -0.3632 |
| 19 | 1.74 | 1.383 | 0.3573 |
| 20 | 2.36 | 1.456 | 0.9045 |
| 21 | 0.91 | 2.046 | -1.136 |
| 22 | 3.24 | 4.477 | -1.237 |
| 23 | 2.08 | 1.734 | 0.346 |
| 24 | 0.12 | 0.03385 | 0.08615 |
| 25 | 0.04 | 0.005131 | 0.03487 |
| 26 | 0.19 | 0.4284 | -0.2384 |
| 27 | 5 | 5.051 | -0.0505 |
| 28 | 0.08 | -0.01427 | 0.09427 |
| 29 | 0.01 | 0.06901 | -0.05901 |
| 30 | 2.04 | 2.34 | -0.2997 |
| 31 | 2.32 | 1.573 | 0.7465 |
| 32 | 0.67 | 1.53 | -0.8599 |
| 33 | 0.25 | 0.3939 | -0.1439 |
| 34 | 0.47 | 0.732 | -0.262 |
| 35 | 0.07 | 0.1235 | -0.05353 |
| 36 | 1.37 | 1.751 | -0.3811 |
| 37 | 2.21 | 2.302 | -0.09242 |
| 38 | 1.23 | 1.538 | -0.308 |
| 39 | 2.94 | 3.468 | -0.5284 |
| 40 | 3.42 | 2.919 | 0.5014 |
| 41 | 2.6 | 5.624 | -3.024 |
| 42 | 1.47 | 1.5 | -0.0295 |
| 43 | 0.86 | 1.403 | -0.5428 |
| 44 | 1.08 | 1.417 | -0.3374 |
| 45 | 1.02 | 1.146 | -0.1259 |
| 46 | 0.84 | 1.09 | -0.2497 |
| 47 | 3.17 | 2.148 | 1.022 |
| 48 | 0.03 | 0.01916 | 0.01084 |
| 49 | 0.07 | 0.151 | -0.08099 |
| 50 | 1.06 | 1.321 | -0.261 |
| 51 | 2.71 | 4.508 | -1.798 |
| 52 | 0.43 | 1.757 | -1.327 |
| 53 | 0.21 | 0.1811 | 0.02886 |
| 54 | 0.83 | 1.381 | -0.5513 |
| 55 | 3.28 | 4.659 | -1.379 |
| 56 | 0.43 | 0.6746 | -0.2446 |
| 57 | 2.58 | 3.158 | -0.5781 |
| 58 | 0.7 | 0.9346 | -0.2346 |
| 59 | 0.16 | 0.08845 | 0.07155 |
| 60 | 0.09 | 0.1374 | -0.04741 |
| 61 | 1.25 | 1.24 | 0.009714 |
| 62 | 0.15 | 0.3236 | -0.1736 |
| 63 | 0.6 | 0.8434 | -0.2434 |
| 64 | 1.9 | 2.265 | -0.3648 |
| 65 | 0.61 | 0.9076 | -0.2976 |
| 66 | 0.64 | 1.185 | -0.5454 |
| 67 | 1.72 | 1.664 | 0.05649 |
| 68 | 1.36 | 1.197 | 0.1631 |
| 69 | 3.22 | 5.041 | -1.821 |
| 70 | 4.59 | 3.742 | 0.848 |
| 71 | 2.77 | 3.987 | -1.217 |
| 72 | 1.09 | 1.379 | -0.2885 |
| 73 | 3.69 | 4.555 | -0.8647 |
| 74 | 1.09 | 1.435 | -0.3453 |
| 75 | 4.59 | 1.971 | 2.619 |
| 76 | 0.2 | 0.541 | -0.341 |
| 77 | 4.17 | 3.138 | 1.032 |
| 78 | 6.89 | 4.036 | 2.854 |
| 79 | 0.95 | 0.8958 | 0.0542 |
| 80 | 0.09 | 0.6164 | -0.5264 |
| 81 | 1.66 | 2.248 | -0.5877 |
| 82 | 2.52 | 1.757 | 0.7634 |
| 83 | 0.51 | 0.3624 | 0.1476 |
| 84 | 0.14 | 0.1177 | 0.02231 |
| 85 | 2.33 | 1.485 | 0.8454 |
| 86 | 2.15 | 2.337 | -0.1868 |
| 87 | 12.65 | 8.912 | 3.738 |
| 88 | 2.06 | 1.444 | 0.6162 |
| 89 | 0.07 | 0.4124 | -0.3424 |
| 90 | 0.07 | 0.1494 | -0.07938 |
| 91 | 2.1 | 1.871 | 0.229 |
| 92 | 0.1 | 0.1047 | -0.004748 |
| 93 | 0.55 | 0.454 | 0.09602 |
| 94 | 1.99 | 1.769 | 0.2214 |
| 95 | 1.74 | 1.805 | -0.06515 |
| 96 | 1.03 | 1.044 | -0.0142 |
| 97 | 2.09 | 1.315 | 0.7752 |
| 98 | 2.13 | 1.776 | 0.3543 |
| 99 | 0.67 | 0.9302 | -0.2602 |
| 100 | 0.17 | 0.09509 | 0.07491 |
| 101 | 0.09 | 0.5192 | -0.4292 |
| 102 | 1.02 | 1.127 | -0.1065 |
| 103 | 0.16 | 0.5163 | -0.3563 |
| 104 | 3.23 | 5.111 | -1.881 |
| 105 | 2.84 | 4.087 | -1.247 |
| 106 | 0.45 | 0.8683 | -0.4183 |
| 107 | 0.1 | -0.1419 | 0.2419 |
| 108 | 0.21 | 0.5278 | -0.3178 |
| 109 | 5.8 | 2.719 | 3.081 |
| 110 | 0.38 | 0.5349 | -0.1549 |
| 111 | 1.44 | 1.753 | -0.3126 |
| 112 | 0.35 | 0.4742 | -0.1242 |
| 113 | 0.97 | 1.154 | -0.1842 |
| 114 | 0.67 | 1.458 | -0.7879 |
| 115 | 0.34 | 1.016 | -0.6764 |
| 116 | 2.64 | 2.32 | 0.3204 |
| 117 | 2.15 | 2.885 | -0.7347 |
| 118 | 9.57 | 7.81 | 1.76 |
| 119 | 1.46 | 1.868 | -0.4076 |
| 120 | 3.87 | 2.163 | 1.707 |
| 121 | 0.07 | 0.3172 | -0.2472 |
| 122 | 3.34 | 1.936 | 1.404 |
| 123 | 1.56 | 1.57 | -0.01013 |
| 124 | 0.96 | 1.246 | -0.2864 |
| 125 | 0.37 | 0.5899 | -0.2199 |
| 126 | 4.21 | 2.907 | 1.303 |
| 127 | 0.3 | 0.2843 | 0.01573 |
| 128 | 1.66 | 1.588 | 0.07242 |
| 129 | 0.07 | 0.05479 | 0.01521 |
| 130 | 5.91 | 5.043 | 0.8672 |
| 131 | 2.82 | 2.595 | 0.2252 |
| 132 | 4.27 | 3.164 | 1.106 |
| 133 | 0 | 0.461 | -0.461 |
| 134 | 2.34 | 1.378 | 0.962 |
| 135 | 2.22 | 3.577 | -1.357 |
| 136 | 0.52 | 1.327 | -0.8073 |
| 137 | 3.01 | 1.905 | 1.105 |
| 138 | 0.67 | 0.7348 | -0.06479 |
| 139 | 3.88 | 5.455 | -1.575 |
| 140 | 4.26 | 7.37 | -3.11 |
| 141 | 0.13 | 0.7788 | -0.6488 |
| 142 | 0.17 | 0.4288 | -0.2588 |
| 143 | 1.54 | 1.406 | 0.1341 |
| 144 | 0.06 | 0.9955 | -0.9355 |
| 145 | 0.31 | 0.2815 | 0.0285 |
| 146 | 0.88 | 1.325 | -0.4454 |
| 147 | 6.89 | 2.378 | 4.512 |
| 148 | 1.11 | 1.323 | -0.2128 |
| 149 | 1.92 | 1.853 | 0.06744 |
| 150 | 4.13 | 1.264 | 2.866 |
| 151 | 0.08 | 0.3187 | -0.2387 |
| 152 | 1.92 | 1.351 | 0.5689 |
| 153 | 3.14 | 4.248 | -1.108 |
| 154 | 6.37 | 4.004 | 2.366 |
| 155 | 5.9 | 4.88 | 1.02 |
| 156 | 0.98 | 2.219 | -1.239 |
| 157 | 1.41 | 0.9895 | 0.4205 |
| 158 | 2.13 | 1.852 | 0.278 |
| 159 | 0.79 | 0.9636 | -0.1736 |
| 160 | 0.42 | 0.4257 | -0.005688 |
| 161 | 0.24 | 0.7161 | -0.4761 |
| 162 | 0.53 | 0.365 | 0.165 |
| Goldfeld-Quandt test for Heteroskedasticity | |||
| p-values | Alternative Hypothesis | ||
| breakpoint index | greater | 2-sided | less |
| 8 | 0.01925 | 0.03851 | 0.9807 |
| 9 | 0.004609 | 0.009219 | 0.9954 |
| 10 | 0.328 | 0.6559 | 0.672 |
| 11 | 0.9355 | 0.129 | 0.0645 |
| 12 | 0.9588 | 0.0824 | 0.0412 |
| 13 | 0.9385 | 0.123 | 0.0615 |
| 14 | 0.9154 | 0.1692 | 0.0846 |
| 15 | 0.8849 | 0.2303 | 0.1151 |
| 16 | 0.837 | 0.326 | 0.163 |
| 17 | 0.7818 | 0.4364 | 0.2182 |
| 18 | 0.7184 | 0.5632 | 0.2816 |
| 19 | 0.6478 | 0.7044 | 0.3522 |
| 20 | 0.6016 | 0.7968 | 0.3984 |
| 21 | 0.5408 | 0.9183 | 0.4592 |
| 22 | 0.5664 | 0.8672 | 0.4336 |
| 23 | 0.4951 | 0.9903 | 0.5049 |
| 24 | 0.4267 | 0.8535 | 0.5733 |
| 25 | 0.359 | 0.718 | 0.641 |
| 26 | 0.2996 | 0.5992 | 0.7004 |
| 27 | 0.2633 | 0.5267 | 0.7367 |
| 28 | 0.2151 | 0.4301 | 0.7849 |
| 29 | 0.1697 | 0.3394 | 0.8303 |
| 30 | 0.1406 | 0.2811 | 0.8594 |
| 31 | 0.2001 | 0.4002 | 0.7999 |
| 32 | 0.1949 | 0.3898 | 0.8051 |
| 33 | 0.1561 | 0.3121 | 0.8439 |
| 34 | 0.1224 | 0.2448 | 0.8776 |
| 35 | 0.09425 | 0.1885 | 0.9057 |
| 36 | 0.07686 | 0.1537 | 0.9231 |
| 37 | 0.05797 | 0.1159 | 0.942 |
| 38 | 0.04556 | 0.09112 | 0.9544 |
| 39 | 0.03643 | 0.07287 | 0.9636 |
| 40 | 0.02854 | 0.05707 | 0.9715 |
| 41 | 0.1373 | 0.2746 | 0.8627 |
| 42 | 0.1093 | 0.2185 | 0.8907 |
| 43 | 0.09423 | 0.1885 | 0.9058 |
| 44 | 0.07652 | 0.153 | 0.9235 |
| 45 | 0.05958 | 0.1192 | 0.9404 |
| 46 | 0.04624 | 0.09248 | 0.9538 |
| 47 | 0.0585 | 0.117 | 0.9415 |
| 48 | 0.04451 | 0.08902 | 0.9555 |
| 49 | 0.03347 | 0.06694 | 0.9665 |
| 50 | 0.02547 | 0.05095 | 0.9745 |
| 51 | 0.03537 | 0.07075 | 0.9646 |
| 52 | 0.03548 | 0.07096 | 0.9645 |
| 53 | 0.02652 | 0.05305 | 0.9735 |
| 54 | 0.02204 | 0.04408 | 0.978 |
| 55 | 0.02293 | 0.04586 | 0.9771 |
| 56 | 0.01725 | 0.0345 | 0.9828 |
| 57 | 0.01339 | 0.02678 | 0.9866 |
| 58 | 0.009845 | 0.01969 | 0.9902 |
| 59 | 0.007024 | 0.01405 | 0.993 |
| 60 | 0.00493 | 0.00986 | 0.9951 |
| 61 | 0.003976 | 0.007952 | 0.996 |
| 62 | 0.002765 | 0.00553 | 0.9972 |
| 63 | 0.001927 | 0.003854 | 0.9981 |
| 64 | 0.001347 | 0.002693 | 0.9987 |
| 65 | 0.001006 | 0.002012 | 0.999 |
| 66 | 0.000744 | 0.001488 | 0.9993 |
| 67 | 0.0004884 | 0.0009768 | 0.9995 |
| 68 | 0.0003217 | 0.0006434 | 0.9997 |
| 69 | 0.0005293 | 0.001059 | 0.9995 |
| 70 | 0.0006924 | 0.001385 | 0.9993 |
| 71 | 0.000693 | 0.001386 | 0.9993 |
| 72 | 0.0004768 | 0.0009536 | 0.9995 |
| 73 | 0.0003836 | 0.0007673 | 0.9996 |
| 74 | 0.0002667 | 0.0005334 | 0.9997 |
| 75 | 0.003672 | 0.007345 | 0.9963 |
| 76 | 0.002685 | 0.00537 | 0.9973 |
| 77 | 0.003057 | 0.006113 | 0.9969 |
| 78 | 0.0436 | 0.08719 | 0.9564 |
| 79 | 0.0339 | 0.06779 | 0.9661 |
| 80 | 0.02806 | 0.05612 | 0.9719 |
| 81 | 0.02364 | 0.04729 | 0.9764 |
| 82 | 0.02059 | 0.04118 | 0.9794 |
| 83 | 0.01561 | 0.03122 | 0.9844 |
| 84 | 0.01163 | 0.02325 | 0.9884 |
| 85 | 0.01025 | 0.02051 | 0.9897 |
| 86 | 0.007627 | 0.01525 | 0.9924 |
| 87 | 0.1813 | 0.3626 | 0.8187 |
| 88 | 0.1623 | 0.3246 | 0.8377 |
| 89 | 0.1382 | 0.2763 | 0.8618 |
| 90 | 0.1145 | 0.2289 | 0.8855 |
| 91 | 0.09478 | 0.1896 | 0.9052 |
| 92 | 0.0768 | 0.1536 | 0.9232 |
| 93 | 0.0616 | 0.1232 | 0.9384 |
| 94 | 0.04932 | 0.09864 | 0.9507 |
| 95 | 0.0389 | 0.07779 | 0.9611 |
| 96 | 0.03014 | 0.06027 | 0.9699 |
| 97 | 0.02715 | 0.0543 | 0.9729 |
| 98 | 0.02131 | 0.04262 | 0.9787 |
| 99 | 0.01634 | 0.03268 | 0.9837 |
| 100 | 0.01218 | 0.02435 | 0.9878 |
| 101 | 0.009417 | 0.01883 | 0.9906 |
| 102 | 0.006858 | 0.01372 | 0.9931 |
| 103 | 0.005107 | 0.01021 | 0.9949 |
| 104 | 0.01026 | 0.02052 | 0.9897 |
| 105 | 0.01207 | 0.02415 | 0.9879 |
| 106 | 0.009337 | 0.01867 | 0.9907 |
| 107 | 0.006995 | 0.01399 | 0.993 |
| 108 | 0.005145 | 0.01029 | 0.9949 |
| 109 | 0.04056 | 0.08113 | 0.9594 |
| 110 | 0.03173 | 0.06346 | 0.9683 |
| 111 | 0.02488 | 0.04976 | 0.9751 |
| 112 | 0.0186 | 0.03721 | 0.9814 |
| 113 | 0.01397 | 0.02794 | 0.986 |
| 114 | 0.01259 | 0.02519 | 0.9874 |
| 115 | 0.01109 | 0.02219 | 0.9889 |
| 116 | 0.008119 | 0.01624 | 0.9919 |
| 117 | 0.007068 | 0.01414 | 0.9929 |
| 118 | 0.02358 | 0.04716 | 0.9764 |
| 119 | 0.02041 | 0.04083 | 0.9796 |
| 120 | 0.02424 | 0.04849 | 0.9758 |
| 121 | 0.01788 | 0.03576 | 0.9821 |
| 122 | 0.02016 | 0.04032 | 0.9798 |
| 123 | 0.01483 | 0.02966 | 0.9852 |
| 124 | 0.01146 | 0.02291 | 0.9885 |
| 125 | 0.008133 | 0.01627 | 0.9919 |
| 126 | 0.00845 | 0.0169 | 0.9916 |
| 127 | 0.005838 | 0.01168 | 0.9942 |
| 128 | 0.004069 | 0.008137 | 0.9959 |
| 129 | 0.002801 | 0.005602 | 0.9972 |
| 130 | 0.003126 | 0.006252 | 0.9969 |
| 131 | 0.002038 | 0.004075 | 0.998 |
| 132 | 0.001875 | 0.003749 | 0.9981 |
| 133 | 0.001233 | 0.002465 | 0.9988 |
| 134 | 0.0009777 | 0.001955 | 0.999 |
| 135 | 0.001155 | 0.002311 | 0.9988 |
| 136 | 0.001289 | 0.002577 | 0.9987 |
| 137 | 0.002021 | 0.004042 | 0.998 |
| 138 | 0.001229 | 0.002459 | 0.9988 |
| 139 | 0.0009996 | 0.001999 | 0.999 |
| 140 | 0.01694 | 0.03388 | 0.9831 |
| 141 | 0.01433 | 0.02867 | 0.9857 |
| 142 | 0.009146 | 0.01829 | 0.9909 |
| 143 | 0.00567 | 0.01134 | 0.9943 |
| 144 | 0.005541 | 0.01108 | 0.9945 |
| 145 | 0.003237 | 0.006474 | 0.9968 |
| 146 | 0.002724 | 0.005449 | 0.9973 |
| 147 | 0.1771 | 0.3541 | 0.8229 |
| 148 | 0.1391 | 0.2781 | 0.8609 |
| 149 | 0.09864 | 0.1973 | 0.9014 |
| 150 | 0.5239 | 0.9523 | 0.4761 |
| 151 | 0.4168 | 0.8336 | 0.5832 |
| 152 | 0.3457 | 0.6914 | 0.6543 |
| 153 | 0.9975 | 0.005039 | 0.00252 |
| 154 | 0.9886 | 0.02275 | 0.01137 |
| Meta Analysis of Goldfeld-Quandt test for Heteroskedasticity | |||
| Description | # significant tests | % significant tests | OK/NOK |
| 1% type I error level | 34 | 0.2313 | NOK |
| 5% type I error level | 81 | 0.55102 | NOK |
| 10% type I error level | 101 | 0.687075 | NOK |
| Ramsey RESET F-Test for powers (2 and 3) of fitted values |
> reset_test_fitted RESET test data: mylm RESET = 27.945, df1 = 2, df2 = 155, p-value = 4.328e-11 |
| Ramsey RESET F-Test for powers (2 and 3) of regressors |
> reset_test_regressors RESET test data: mylm RESET = 11.408, df1 = 8, df2 = 149, p-value = 1.519e-12 |
| Ramsey RESET F-Test for powers (2 and 3) of principal components |
> reset_test_principal_components RESET test data: mylm RESET = 32.554, df1 = 2, df2 = 155, p-value = 1.572e-12 |
| Variance Inflation Factors (Multicollinearity) |
> vif
`Population_(millions)` HDI GDP_per_Capita
1.007334 1.857608 1.855642
Total_Biocapacity
1.007501
|