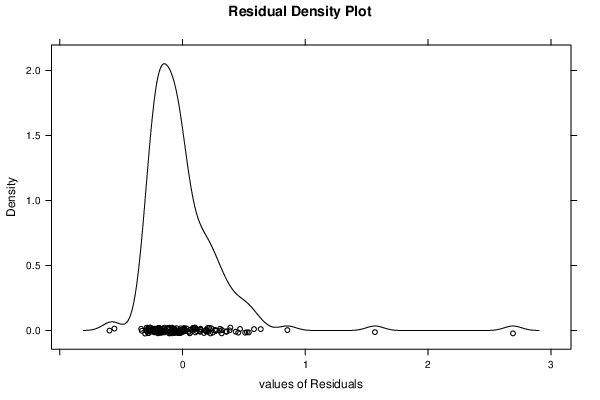
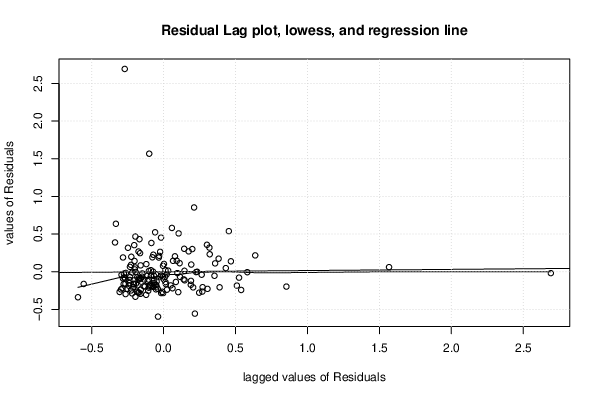
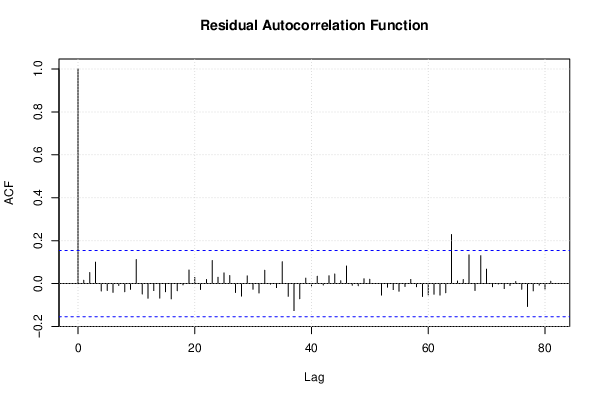
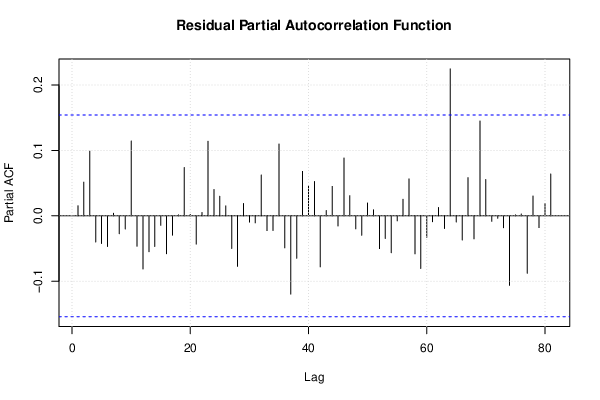
| Multiple Linear Regression - Estimated Regression Equation |
| Forest_Footprint[t] = + 0.1978 -0.000144007`Population_(millions)`[t] + 0.14764HDI[t] + 4.14275e-06GDP_per_Capita[t] + 0.00826966Total_Biocapacity[t] + e[t] |
| Multiple Linear Regression - Ordinary Least Squares | |||||
| Variable | Parameter | S.D. | T-STAT H0: parameter = 0 | 2-tail p-value | 1-tail p-value |
| (Intercept) | +0.1978 | 0.1465 | +1.3500e+00 | 0.1789 | 0.08947 |
| `Population_(millions)` | -0.000144 | 0.0001788 | -8.0560e-01 | 0.4217 | 0.2109 |
| HDI | +0.1476 | 0.2341 | +6.3050e-01 | 0.5293 | 0.2646 |
| GDP_per_Capita | +4.143e-06 | 1.858e-06 | +2.2300e+00 | 0.02719 | 0.0136 |
| Total_Biocapacity | +0.00827 | 0.002933 | +2.8190e+00 | 0.005434 | 0.002717 |
| Multiple Linear Regression - Regression Statistics | |
| Multiple R | 0.3595 |
| R-squared | 0.1292 |
| Adjusted R-squared | 0.107 |
| F-TEST (value) | 5.825 |
| F-TEST (DF numerator) | 4 |
| F-TEST (DF denominator) | 157 |
| p-value | 0.0002137 |
| Multiple Linear Regression - Residual Statistics | |
| Residual Standard Deviation | 0.3412 |
| Sum Squared Residuals | 18.28 |
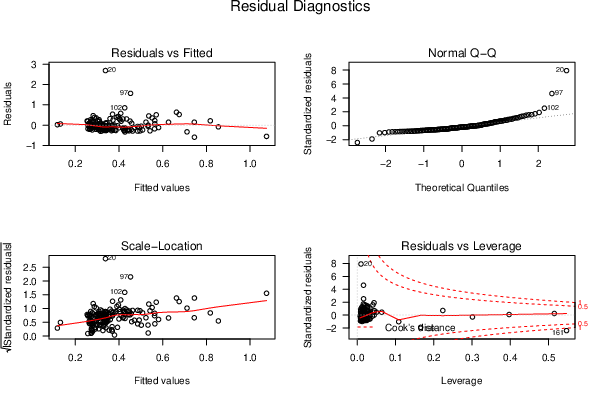
| Menu of Residual Diagnostics | |
| Description | Link |
| Histogram | Compute |
| Central Tendency | Compute |
| QQ Plot | Compute |
| Kernel Density Plot | Compute |
| Skewness/Kurtosis Test | Compute |
| Skewness-Kurtosis Plot | Compute |
| Harrell-Davis Plot | Compute |
| Bootstrap Plot -- Central Tendency | Compute |
| Blocked Bootstrap Plot -- Central Tendency | Compute |
| (Partial) Autocorrelation Plot | Compute |
| Spectral Analysis | Compute |
| Tukey lambda PPCC Plot | Compute |
| Box-Cox Normality Plot | Compute |
| Summary Statistics | Compute |
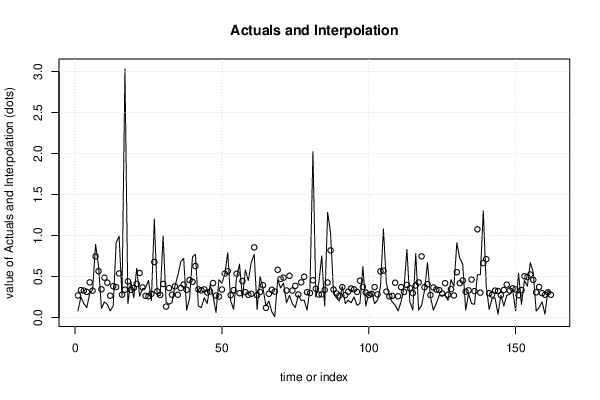
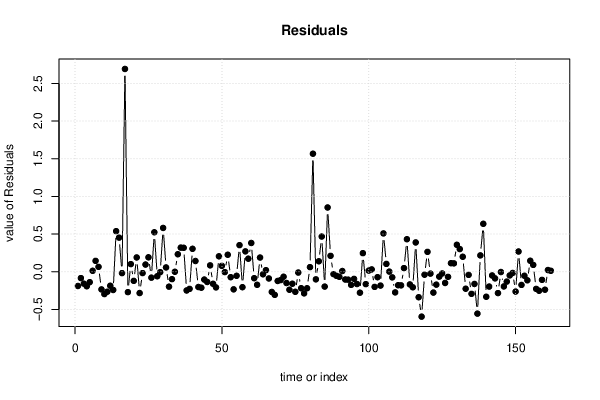
| Multiple Linear Regression - Actuals, Interpolation, and Residuals | |||
| Time or Index | Actuals | Interpolation Forecast | Residuals Prediction Error |
| 1 | 0.08 | 0.2681 | -0.1881 |
| 2 | 0.25 | 0.3337 | -0.08367 |
| 3 | 0.17 | 0.3274 | -0.1574 |
| 4 | 0.12 | 0.312 | -0.192 |
| 5 | 0.29 | 0.4277 | -0.1377 |
| 6 | 0.34 | 0.3267 | 0.0133 |
| 7 | 0.89 | 0.7447 | 0.1453 |
| 8 | 0.63 | 0.5643 | 0.06569 |
| 9 | 0.11 | 0.3437 | -0.2337 |
| 10 | 0.19 | 0.4857 | -0.2957 |
| 11 | 0.16 | 0.4241 | -0.2641 |
| 12 | 0.08 | 0.2649 | -0.1849 |
| 13 | 0.14 | 0.3811 | -0.2411 |
| 14 | 0.91 | 0.3708 | 0.5392 |
| 15 | 0.99 | 0.5365 | 0.4535 |
| 16 | 0.26 | 0.2776 | -0.01759 |
| 17 | 3.03 | 0.3383 | 2.692 |
| 18 | 0.17 | 0.4399 | -0.2699 |
| 19 | 0.44 | 0.338 | 0.102 |
| 20 | 0.24 | 0.3602 | -0.1202 |
| 21 | 0.6 | 0.4098 | 0.1902 |
| 22 | 0.26 | 0.5421 | -0.2821 |
| 23 | 0.35 | 0.3671 | -0.01711 |
| 24 | 0.36 | 0.2639 | 0.09611 |
| 25 | 0.45 | 0.2578 | 0.1922 |
| 26 | 0.21 | 0.2877 | -0.07774 |
| 27 | 1.2 | 0.6756 | 0.5244 |
| 28 | 0.26 | 0.3189 | -0.05891 |
| 29 | 0.27 | 0.2752 | -0.005185 |
| 30 | 0.99 | 0.408 | 0.582 |
| 31 | 0.19 | 0.1321 | 0.05786 |
| 32 | 0.16 | 0.3573 | -0.1973 |
| 33 | 0.18 | 0.2777 | -0.09773 |
| 34 | 0.38 | 0.3796 | 0.0003956 |
| 35 | 0.51 | 0.2771 | 0.2329 |
| 36 | 0.68 | 0.3592 | 0.3208 |
| 37 | 0.72 | 0.4016 | 0.3184 |
| 38 | 0.09 | 0.3376 | -0.2476 |
| 39 | 0.23 | 0.4563 | -0.2263 |
| 40 | 0.74 | 0.4349 | 0.3051 |
| 41 | 0.77 | 0.6268 | 0.1432 |
| 42 | 0.14 | 0.342 | -0.202 |
| 43 | 0.12 | 0.331 | -0.211 |
| 44 | 0.24 | 0.3431 | -0.1031 |
| 45 | 0.17 | 0.3048 | -0.1348 |
| 46 | 0.4 | 0.3147 | 0.08531 |
| 47 | 0.26 | 0.4194 | -0.1594 |
| 48 | 0.06 | 0.2671 | -0.2071 |
| 49 | 0.46 | 0.2544 | 0.2056 |
| 50 | 0.42 | 0.341 | 0.07902 |
| 51 | 0.53 | 0.5339 | -0.003911 |
| 52 | 0.79 | 0.5637 | 0.2263 |
| 53 | 0.2 | 0.2714 | -0.07138 |
| 54 | 0.1 | 0.333 | -0.233 |
| 55 | 0.48 | 0.533 | -0.05297 |
| 56 | 0.65 | 0.2962 | 0.3538 |
| 57 | 0.24 | 0.4441 | -0.2041 |
| 58 | 0.58 | 0.3087 | 0.2713 |
| 59 | 0.45 | 0.2759 | 0.1741 |
| 60 | 0.67 | 0.2874 | 0.3826 |
| 61 | 0.77 | 0.8548 | -0.08484 |
| 62 | 0.1 | 0.2725 | -0.1725 |
| 63 | 0.5 | 0.3108 | 0.1892 |
| 64 | 0.36 | 0.3932 | -0.03322 |
| 65 | 0.14 | 0.1183 | 0.02172 |
| 66 | 0.2 | 0.2883 | -0.08835 |
| 67 | 0.07 | 0.3376 | -0.2676 |
| 68 | 0.01 | 0.3157 | -0.3057 |
| 69 | 0.46 | 0.5813 | -0.1213 |
| 70 | 0.36 | 0.4707 | -0.1107 |
| 71 | 0.42 | 0.4855 | -0.06554 |
| 72 | 0.18 | 0.3289 | -0.1489 |
| 73 | 0.27 | 0.5082 | -0.2382 |
| 74 | 0.17 | 0.3284 | -0.1584 |
| 75 | 0.12 | 0.3855 | -0.2655 |
| 76 | 0.27 | 0.2799 | -0.009925 |
| 77 | 0.21 | 0.4278 | -0.2178 |
| 78 | 0.21 | 0.4962 | -0.2862 |
| 79 | 0.09 | 0.3084 | -0.2184 |
| 80 | 0.36 | 0.298 | 0.06196 |
| 81 | 2.02 | 0.453 | 1.567 |
| 82 | 0.25 | 0.3499 | -0.09995 |
| 83 | 0.42 | 0.2804 | 0.1396 |
| 84 | 0.75 | 0.2821 | 0.4679 |
| 85 | 0.14 | 0.3362 | -0.1962 |
| 86 | 1.28 | 0.4263 | 0.8537 |
| 87 | 1.03 | 0.818 | 0.212 |
| 88 | 0.31 | 0.3407 | -0.03068 |
| 89 | 0.24 | 0.2935 | -0.05353 |
| 90 | 0.2 | 0.2665 | -0.0665 |
| 91 | 0.38 | 0.3697 | 0.01032 |
| 92 | 0.17 | 0.2723 | -0.1023 |
| 93 | 0.21 | 0.3144 | -0.1044 |
| 94 | 0.18 | 0.3537 | -0.1737 |
| 95 | 0.25 | 0.3436 | -0.09357 |
| 96 | 0.15 | 0.3125 | -0.1625 |
| 97 | 0.17 | 0.4472 | -0.2772 |
| 98 | 0.62 | 0.3727 | 0.2473 |
| 99 | 0.14 | 0.3036 | -0.1636 |
| 100 | 0.29 | 0.274 | 0.01603 |
| 101 | 0.32 | 0.2883 | 0.03171 |
| 102 | 0.17 | 0.3703 | -0.2003 |
| 103 | 0.21 | 0.2813 | -0.07135 |
| 104 | 0.38 | 0.5629 | -0.1829 |
| 105 | 1.08 | 0.5707 | 0.5093 |
| 106 | 0.42 | 0.3153 | 0.1047 |
| 107 | 0.26 | 0.2575 | 0.002517 |
| 108 | 0.19 | 0.2639 | -0.07392 |
| 109 | 0.15 | 0.4236 | -0.2736 |
| 110 | 0.08 | 0.2582 | -0.1782 |
| 111 | 0.19 | 0.3701 | -0.1801 |
| 112 | 0.36 | 0.3108 | 0.04924 |
| 113 | 0.83 | 0.3975 | 0.4325 |
| 114 | 0.19 | 0.3576 | -0.1676 |
| 115 | 0.09 | 0.2956 | -0.2056 |
| 116 | 0.78 | 0.3906 | 0.3894 |
| 117 | 0.09 | 0.4275 | -0.3375 |
| 118 | 0.15 | 0.7452 | -0.5952 |
| 119 | 0.33 | 0.3687 | -0.03866 |
| 120 | 0.67 | 0.4052 | 0.2648 |
| 121 | 0.25 | 0.2741 | -0.02408 |
| 122 | 0.09 | 0.3658 | -0.2758 |
| 123 | 0.17 | 0.3404 | -0.1704 |
| 124 | 0.27 | 0.3339 | -0.06393 |
| 125 | 0.27 | 0.2919 | -0.02191 |
| 126 | 0.27 | 0.4181 | -0.1481 |
| 127 | 0.21 | 0.2769 | -0.06689 |
| 128 | 0.46 | 0.3456 | 0.1144 |
| 129 | 0.38 | 0.2683 | 0.1117 |
| 130 | 0.91 | 0.5519 | 0.3581 |
| 131 | 0.72 | 0.4184 | 0.3016 |
| 132 | 0.65 | 0.4506 | 0.1994 |
| 133 | 0.09 | 0.3144 | -0.2244 |
| 134 | 0.29 | 0.3307 | -0.04072 |
| 135 | 0.17 | 0.4625 | -0.2925 |
| 136 | 0.16 | 0.3211 | -0.1611 |
| 137 | 0.52 | 1.075 | -0.5552 |
| 138 | 0.52 | 0.3024 | 0.2176 |
| 139 | 1.3 | 0.6631 | 0.6369 |
| 140 | 0.38 | 0.7114 | -0.3314 |
| 141 | 0.1 | 0.296 | -0.196 |
| 142 | 0.23 | 0.2783 | -0.04832 |
| 143 | 0.24 | 0.3274 | -0.08744 |
| 144 | 0.04 | 0.3224 | -0.2824 |
| 145 | 0.27 | 0.273 | -0.003024 |
| 146 | 0.14 | 0.3343 | -0.1943 |
| 147 | 0.27 | 0.4 | -0.13 |
| 148 | 0.28 | 0.3281 | -0.04806 |
| 149 | 0.34 | 0.3552 | -0.01516 |
| 150 | 0.08 | 0.3424 | -0.2624 |
| 151 | 0.54 | 0.2708 | 0.2692 |
| 152 | 0.16 | 0.3341 | -0.1741 |
| 153 | 0.45 | 0.5023 | -0.05229 |
| 154 | 0.38 | 0.4927 | -0.1127 |
| 155 | 0.67 | 0.5235 | 0.1465 |
| 156 | 0.55 | 0.4583 | 0.09173 |
| 157 | 0.08 | 0.3067 | -0.2267 |
| 158 | 0.12 | 0.3717 | -0.2517 |
| 159 | 0.19 | 0.2968 | -0.1068 |
| 160 | 0.04 | 0.2777 | -0.2377 |
| 161 | 0.33 | 0.3071 | 0.02294 |
| 162 | 0.29 | 0.2769 | 0.01312 |
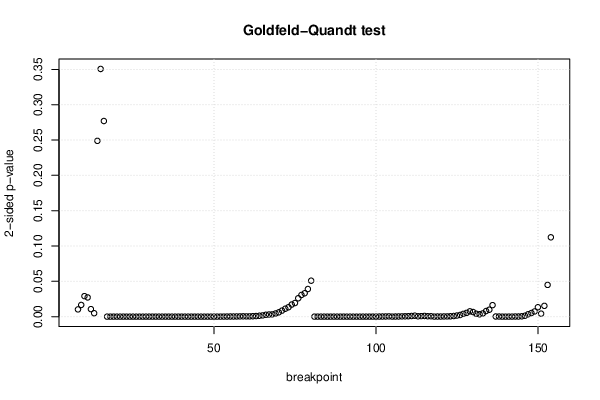
| Goldfeld-Quandt test for Heteroskedasticity | |||
| p-values | Alternative Hypothesis | ||
| breakpoint index | greater | 2-sided | less |
| 8 | 0.005119 | 0.01024 | 0.9949 |
| 9 | 0.008285 | 0.01657 | 0.9917 |
| 10 | 0.01452 | 0.02904 | 0.9855 |
| 11 | 0.01361 | 0.02721 | 0.9864 |
| 12 | 0.005336 | 0.01067 | 0.9947 |
| 13 | 0.002438 | 0.004876 | 0.9976 |
| 14 | 0.1244 | 0.2489 | 0.8756 |
| 15 | 0.1753 | 0.3507 | 0.8247 |
| 16 | 0.1385 | 0.2769 | 0.8615 |
| 17 | 1 | 3.182e-11 | 1.591e-11 |
| 18 | 1 | 8.944e-12 | 4.472e-12 |
| 19 | 1 | 2.557e-11 | 1.279e-11 |
| 20 | 1 | 5.368e-11 | 2.684e-11 |
| 21 | 1 | 5.953e-11 | 2.977e-11 |
| 22 | 1 | 9.448e-11 | 4.724e-11 |
| 23 | 1 | 2.455e-10 | 1.227e-10 |
| 24 | 1 | 5.686e-10 | 2.843e-10 |
| 25 | 1 | 1.241e-09 | 6.205e-10 |
| 26 | 1 | 2.556e-09 | 1.278e-09 |
| 27 | 1 | 1.962e-09 | 9.811e-10 |
| 28 | 1 | 3.294e-09 | 1.647e-09 |
| 29 | 1 | 7.234e-09 | 3.617e-09 |
| 30 | 1 | 2.974e-09 | 1.487e-09 |
| 31 | 1 | 5.791e-09 | 2.895e-09 |
| 32 | 1 | 9.313e-09 | 4.657e-09 |
| 33 | 1 | 1.88e-08 | 9.401e-09 |
| 34 | 1 | 3.655e-08 | 1.828e-08 |
| 35 | 1 | 5.991e-08 | 2.996e-08 |
| 36 | 1 | 7.561e-08 | 3.781e-08 |
| 37 | 1 | 9.765e-08 | 4.883e-08 |
| 38 | 1 | 1.347e-07 | 6.736e-08 |
| 39 | 1 | 2.06e-07 | 1.03e-07 |
| 40 | 1 | 2.601e-07 | 1.3e-07 |
| 41 | 1 | 4.553e-07 | 2.276e-07 |
| 42 | 1 | 6.908e-07 | 3.454e-07 |
| 43 | 1 | 1.037e-06 | 5.184e-07 |
| 44 | 1 | 1.802e-06 | 9.008e-07 |
| 45 | 1 | 3.027e-06 | 1.514e-06 |
| 46 | 1 | 5.262e-06 | 2.631e-06 |
| 47 | 1 | 7.968e-06 | 3.984e-06 |
| 48 | 1 | 1.153e-05 | 5.767e-06 |
| 49 | 1 | 1.646e-05 | 8.23e-06 |
| 50 | 1 | 2.75e-05 | 1.375e-05 |
| 51 | 1 | 4.611e-05 | 2.306e-05 |
| 52 | 1 | 6.251e-05 | 3.126e-05 |
| 53 | 1 | 0.0001008 | 5.041e-05 |
| 54 | 0.9999 | 0.0001319 | 6.597e-05 |
| 55 | 0.9999 | 0.0002094 | 0.0001047 |
| 56 | 0.9999 | 0.000206 | 0.000103 |
| 57 | 0.9999 | 0.0002808 | 0.0001404 |
| 58 | 0.9998 | 0.0003328 | 0.0001664 |
| 59 | 0.9998 | 0.0004627 | 0.0002314 |
| 60 | 0.9998 | 0.0004166 | 0.0002083 |
| 61 | 0.9998 | 0.0004562 | 0.0002281 |
| 62 | 0.9997 | 0.0006247 | 0.0003124 |
| 63 | 0.9996 | 0.0008348 | 0.0004174 |
| 64 | 0.9994 | 0.001253 | 0.0006266 |
| 65 | 0.9991 | 0.001803 | 0.0009017 |
| 66 | 0.9987 | 0.002603 | 0.001302 |
| 67 | 0.9985 | 0.00304 | 0.00152 |
| 68 | 0.9984 | 0.003288 | 0.001644 |
| 69 | 0.9978 | 0.004477 | 0.002238 |
| 70 | 0.9969 | 0.006135 | 0.003067 |
| 71 | 0.9957 | 0.008554 | 0.004277 |
| 72 | 0.9944 | 0.01121 | 0.005604 |
| 73 | 0.9933 | 0.01335 | 0.006673 |
| 74 | 0.9914 | 0.01711 | 0.008556 |
| 75 | 0.9903 | 0.0193 | 0.009651 |
| 76 | 0.987 | 0.02605 | 0.01303 |
| 77 | 0.9846 | 0.03075 | 0.01538 |
| 78 | 0.9834 | 0.03314 | 0.01657 |
| 79 | 0.9804 | 0.03914 | 0.01957 |
| 80 | 0.9746 | 0.0509 | 0.02545 |
| 81 | 1 | 7.336e-06 | 3.668e-06 |
| 82 | 1 | 1.206e-05 | 6.031e-06 |
| 83 | 1 | 1.765e-05 | 8.826e-06 |
| 84 | 1 | 6.799e-06 | 3.399e-06 |
| 85 | 1 | 9.603e-06 | 4.802e-06 |
| 86 | 1 | 1.214e-07 | 6.07e-08 |
| 87 | 1 | 1.327e-07 | 6.637e-08 |
| 88 | 1 | 2.544e-07 | 1.272e-07 |
| 89 | 1 | 4.738e-07 | 2.369e-07 |
| 90 | 1 | 8.685e-07 | 4.342e-07 |
| 91 | 1 | 1.598e-06 | 7.988e-07 |
| 92 | 1 | 2.826e-06 | 1.413e-06 |
| 93 | 1 | 4.937e-06 | 2.468e-06 |
| 94 | 1 | 7.494e-06 | 3.747e-06 |
| 95 | 1 | 1.249e-05 | 6.246e-06 |
| 96 | 1 | 1.928e-05 | 9.638e-06 |
| 97 | 1 | 2.354e-05 | 1.177e-05 |
| 98 | 1 | 2.715e-05 | 1.358e-05 |
| 99 | 1 | 4.178e-05 | 2.089e-05 |
| 100 | 1 | 6.757e-05 | 3.378e-05 |
| 101 | 0.9999 | 0.00011 | 5.498e-05 |
| 102 | 0.9999 | 0.0001604 | 8.021e-05 |
| 103 | 0.9999 | 0.0002632 | 0.0001316 |
| 104 | 0.9998 | 0.0003786 | 0.0001893 |
| 105 | 0.9999 | 0.0001052 | 5.26e-05 |
| 106 | 0.9999 | 0.0001565 | 7.827e-05 |
| 107 | 0.9999 | 0.0002482 | 0.0001241 |
| 108 | 0.9998 | 0.0004028 | 0.0002014 |
| 109 | 0.9998 | 0.0004874 | 0.0002437 |
| 110 | 0.9997 | 0.0006469 | 0.0003234 |
| 111 | 0.9995 | 0.0009277 | 0.0004639 |
| 112 | 0.9993 | 0.001373 | 0.0006865 |
| 113 | 0.9997 | 0.0005408 | 0.0002704 |
| 114 | 0.9996 | 0.0008019 | 0.000401 |
| 115 | 0.9995 | 0.0009952 | 0.0004976 |
| 116 | 0.9997 | 0.0006136 | 0.0003068 |
| 117 | 0.9997 | 0.0006065 | 0.0003033 |
| 118 | 1 | 9.257e-05 | 4.628e-05 |
| 119 | 0.9999 | 0.000167 | 8.352e-05 |
| 120 | 0.9999 | 0.0001484 | 7.421e-05 |
| 121 | 0.9999 | 0.0002683 | 0.0001342 |
| 122 | 0.9998 | 0.0003232 | 0.0001616 |
| 123 | 0.9997 | 0.0005209 | 0.0002605 |
| 124 | 0.9995 | 0.000914 | 0.000457 |
| 125 | 0.9992 | 0.00157 | 0.0007849 |
| 126 | 0.9988 | 0.002381 | 0.00119 |
| 127 | 0.998 | 0.003979 | 0.00199 |
| 128 | 0.9973 | 0.005384 | 0.002692 |
| 129 | 0.9962 | 0.007544 | 0.003772 |
| 130 | 0.9967 | 0.006518 | 0.003259 |
| 131 | 0.998 | 0.00406 | 0.00203 |
| 132 | 0.9983 | 0.003332 | 0.001666 |
| 133 | 0.9977 | 0.0046 | 0.0023 |
| 134 | 0.996 | 0.007941 | 0.003971 |
| 135 | 0.9951 | 0.009782 | 0.004891 |
| 136 | 0.9918 | 0.01631 | 0.008155 |
| 137 | 0.9999 | 0.0002265 | 0.0001132 |
| 138 | 0.9999 | 0.0001735 | 8.674e-05 |
| 139 | 1 | 1.038e-05 | 5.192e-06 |
| 140 | 1 | 9.492e-06 | 4.746e-06 |
| 141 | 1 | 2.59e-05 | 1.295e-05 |
| 142 | 1 | 7.216e-05 | 3.608e-05 |
| 143 | 0.9999 | 0.0001908 | 9.54e-05 |
| 144 | 0.9999 | 0.0001598 | 7.989e-05 |
| 145 | 0.9998 | 0.0004639 | 0.0002319 |
| 146 | 0.9994 | 0.001286 | 0.0006428 |
| 147 | 0.9983 | 0.00343 | 0.001715 |
| 148 | 0.9974 | 0.005195 | 0.002598 |
| 149 | 0.9963 | 0.007411 | 0.003705 |
| 150 | 0.9933 | 0.01335 | 0.006677 |
| 151 | 0.9979 | 0.004234 | 0.002117 |
| 152 | 0.9923 | 0.0153 | 0.007651 |
| 153 | 0.9775 | 0.04495 | 0.02247 |
| 154 | 0.9438 | 0.1123 | 0.05616 |
| Meta Analysis of Goldfeld-Quandt test for Heteroskedasticity | |||
| Description | # significant tests | % significant tests | OK/NOK |
| 1% type I error level | 125 | 0.8503 | NOK |
| 5% type I error level | 142 | 0.965986 | NOK |
| 10% type I error level | 143 | 0.972789 | NOK |
| Ramsey RESET F-Test for powers (2 and 3) of fitted values |
> reset_test_fitted RESET test data: mylm RESET = 4.7239, df1 = 2, df2 = 155, p-value = 0.0102 |
| Ramsey RESET F-Test for powers (2 and 3) of regressors |
> reset_test_regressors RESET test data: mylm RESET = 4.5707, df1 = 8, df2 = 149, p-value = 5.342e-05 |
| Ramsey RESET F-Test for powers (2 and 3) of principal components |
> reset_test_principal_components RESET test data: mylm RESET = 0.19068, df1 = 2, df2 = 155, p-value = 0.8266 |
| Variance Inflation Factors (Multicollinearity) |
> vif
`Population_(millions)` HDI GDP_per_Capita
1.007334 1.857608 1.855642
Total_Biocapacity
1.007501
|