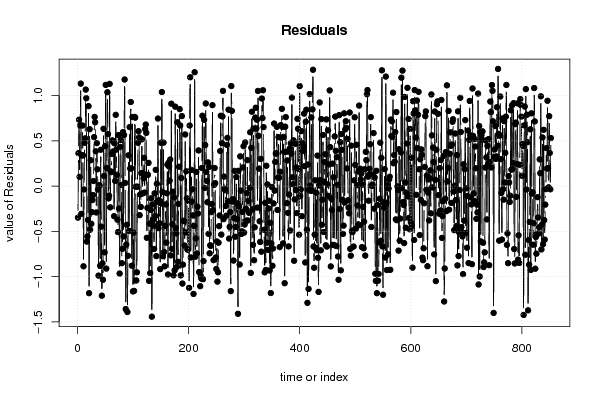
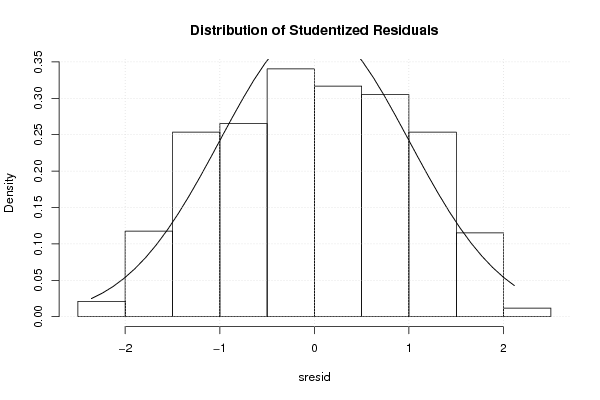
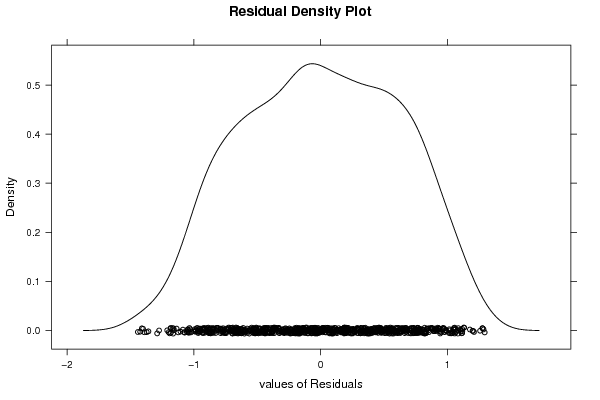
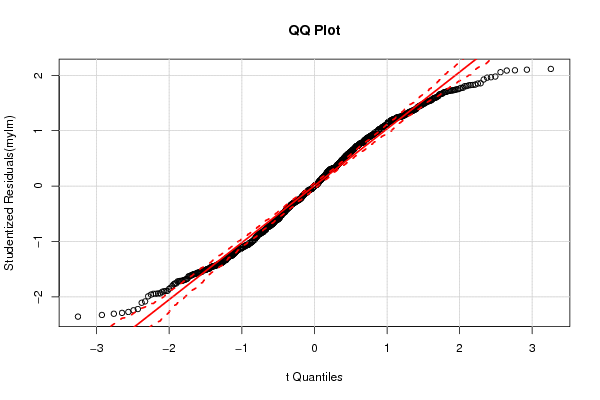
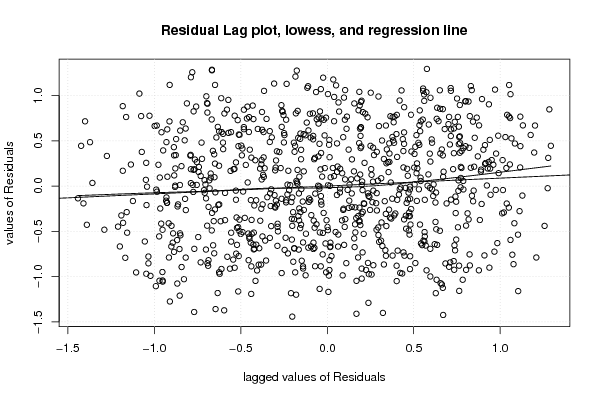
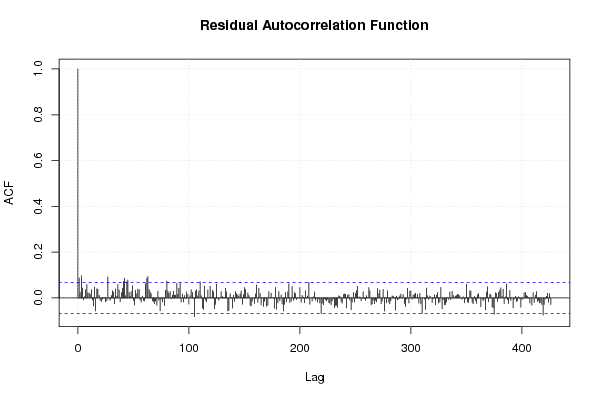
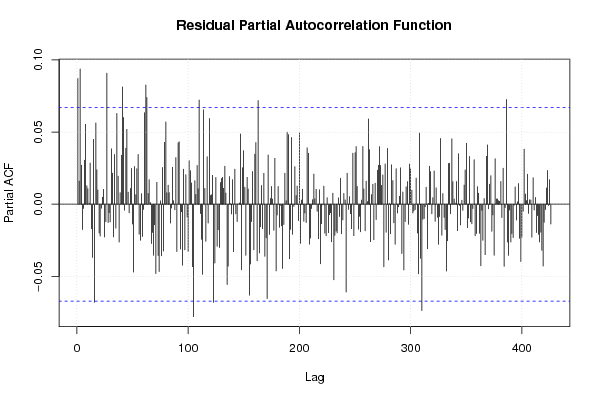
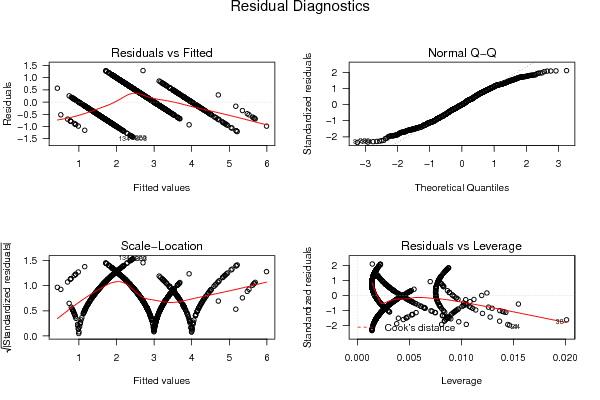
1 5 -0,5270
1 4 -0,0906
1 3 0,2587
1 3 0,0979
1 3 0,2428
1 3 0,3605
1 4 -0,2615
1 3 0,1583
1 3 0,2424
1 3 0,1584
1 1 0,3561
1 4 -0,1927
1 4 -0,0613
1 3 0,2063
1 3 0,3436
1 3 0,3194
1 3 -0,0845
1 1 0,4403
1 4 -0,2787
1 3 0,2970
1 4 -0,4848
1 3 0,2320
1 4 -0,3061
1 3 0,1446
1 1 0,4758
1 3 -0,0013
1 4 -0,2226
1 4 -0,2083
1 3 0,0862
1 3 0,2098
1 3 0,2666
1 3 0,2516
1 3 0,1699
1 3 -0,0024
1 4 -0,0622
1 3 0,1190
1 3 0,0620
1 5 -0,6904
1 3 0,0743
1 3 0,0730
1 4 -0,4075
1 4 -0,0808
1 4 -0,2974
1 4 -0,4918
1 1 0,3644
1 1 0,3182
1 3 0,2332
1 4 -0,1547
1 4 -0,3698
1 3 0,1843
1 3 0,3568
1 1 0,3496
1 3 0,1227
1 3 0,3361
1 3 0,2182
1 3 0,0371
1 4 -0,2099
1 3 0,3597
0 3 -0,0412
0 4 -0,1949
0 3 0,0581
0 4 -0,1459
0 4 -0,1076
0 4 -0,1391
0 4 -0,1122
0 3 -0,0660
0 3 0,1389
0 4 -0,2219
0 3 0,2193
0 4 -0,1337
0 1 0,5604
0 1 0,4342
0 1 0,3993
0 3 0,1623
0 4 -0,2995
0 1 0,2825
0 3 0,1293
0 3 0,1486
0 1 0,5319
0 1 0,3116
0 1 0,3521
0 3 0,1707
0 1 0,3869
0 3 0,1626
0 3 0,3188
0 3 0,0276
0 1 0,1824
0 4 -0,4011
0 1 0,4198
0 1 0,1742
0 1 0,3325
0 3 0,1053
0 1 0,4029
0 3 0,1849
0 3 0,0683
0 3 0,2555
0 3 0,0684
0 0 0,5597
0 3 0,2132
0 1 0,2321
0 3 -0,1126
0 0 0,4887
0 3 0,2134
0 3 0,2114
0 4 -0,2381
0 1 0,2630
0 1 0,2860
0 3 0,0171
0 3 0,1407
0 3 0,1733
0 3 0,1558
0 3 -0,0631
0 3 -0,0048
0 1 0,4701
0 1 0,5651
0 3 0,1504
0 3 -0,0021
0 3 0,0549
0 4 -0,2143
0 3 0,0983
0 3 0,0005
0 3 0,1792
0 3 0,1920
0 3 0,1685
0 1 0,3832
0 3 -0,0427
0 1 0,5606
0 3 0,0841
0 1 0,2618
0 1 0,4230
0 1 0,2839
0 1 0,4616
0 3 -0,0158
0 1 0,1611
0 1 0,4772
0 1 0,4335
0 1 0,4172
0 1 0,3517
0 1 0,5083
0 3 -0,0655
0 3 0,0645
0 0 0,5866
0 1 0,5351
0 1 0,4848
0 3 0,2089
0 1 0,4103
0 3 -0,0212
0 1 0,4240
0 0 0,5500
0 3 0,1408
0 1 0,4796
0 3 0,2835
0 3 0,0022
0 1 0,3312
0 4 -0,1140
0 1 0,3434
0 1 0,3028
0 1 0,4168
0 1 0,4772
0 1 0,3695
0 3 -0,0016
0 3 0,0787
0 1 0,2798
0 1 0,5942
0 4 -0,1813
0 4 -0,2211
0 5 -0,4167
0 1 0,3336
0 3 0,2506
0 1 0,3522
0 1 0,4213
0 1 0,5130
0 1 0,2776
0 1 0,4969
0 1 0,2916
0 3 0,2424
0 1 0,4111
0 0 0,6507
0 4 -0,4637
0 3 0,1984
0 1 0,3157
0 3 -0,0314
0 4 -0,1818
0 3 0,2354
0 3 0,1892
0 0 0,5324
0 1 0,5516
0 3 0,2158
0 1 0,2543
0 1 0,3076
0 1 0,4312
0 1 0,4300
0 1 0,3614
0 1 0,6741
0 1 0,3984
0 4 -0,1949
0 1 0,4942
0 0 0,6051
0 1 0,3499
0 1 0,4868
0 1 0,2422
0 3 0,1892
0 3 0,3252
0 0 0,5828
0 1 0,5743
0 5 -0,5352
0 1 0,5198
0 1 0,4462
0 1 0,2251
0 1 0,4167
0 3 0,3393
0 1 0,3297
0 1 0,3814
0 1 0,5686
0 1 0,3391
0 1 0,4708
0 3 -0,0585
0 3 0,1191
0 1 0,2868
0 1 0,2857
0 1 0,2469
0 1 0,2760
0 1 0,2681
0 1 0,5797
0 4 -0,0383
0 1 0,2666
0 1 0,3175
0 3 0,2053
0 1 0,5237
0 3 0,1341
0 3 0,2514
0 3 0,0586
0 4 -0,2218
0 1 0,4833
0 3 0,0854
0 1 0,3942
0 3 0,0725
0 1 0,3395
0 1 0,3621
0 1 0,3605
0 1 0,4553
0 3 -0,0317
0 3 0,2465
0 1 0,4612
0 1 0,3202
0 1 0,5321
0 3 0,0696
0 1 0,5371
0 0 0,5827
0 0 0,5504
0 1 0,3728
0 1 0,2597
0 1 0,2861
0 3 -0,1408
0 1 0,4160
0 3 -0,0640
0 4 -0,1376
0 3 0,2168
0 1 0,3923
0 3 0,1401
0 3 0,2147
0 3 0,2868
0 1 0,4356
0 4 -0,2097
0 3 0,0112
0 4 -0,2479
0 4 -0,2925
0 1 0,4454
0 3 0,1347
0 3 0,1545
0 3 -0,0241
0 3 0,2339
0 1 0,3809
0 1 0,4141
0 1 0,4555
0 1 0,2330
0 3 0,3004
0 3 0,2305
0 3 -0,0249
0 1 0,4927
0 1 0,3191
0 1 0,4386
0 1 0,5716
0 1 0,3862
0 3 -0,0963
0 3 0,0445
0 1 0,4353
0 3 -0,0300
0 1 0,1692
0 1 0,5719
0 1 0,4714
0 1 0,3080
0 3 0,1023
0 3 0,0058
0 1 0,3940
0 3 -0,1088
0 3 -0,0540
0 4 -0,1839
0 4 -0,1239
0 3 -0,1123
0 3 -0,0896
0 4 -0,1135
0 3 -0,0452
0 3 -0,0786
0 3 -0,0012
0 3 0,0917
0 3 -0,0781
0 3 -0,0227
0 3 -0,0570
0 3 0,0134
0 4 -0,0848
0 1 0,2843
0 3 0,1264
0 3 0,2281
0 3 -0,0471
0 5 -0,6570
0 3 0,1808
0 1 0,3200
0 1 0,4128
0 3 0,2115
0 4 -0,0158
0 3 0,1809
0 3 0,2055
0 1 0,4682
0 3 0,2866
0 4 -0,0950
0 3 0,0678
0 4 -0,3354
0 5 -0,6486
0 3 0,0953
0 1 0,3441
0 3 0,2653
0 3 0,2102
0 3 0,2885
0 3 0,1848
0 3 -0,1043
0 4 -0,4793
0 4 -0,2836
0 4 -0,4726
0 4 -0,1791
0 4 -0,3959
0 4 -0,2317
0 4 -0,4160
0 4 -0,2992
0 4 -0,2776
0 3 -0,2189
0 3 -0,0848
0 4 -0,5375
0 3 -0,1429
0 4 -0,2389
0 1 0,3043
0 5 -0,6671
0 3 -0,0302
0 3 0,1948
0 3 0,0059
0 3 0,0867
0 3 0,1104
0 3 0,1105
0 4 -0,0696
0 3 -0,0484
0 3 0,0423
0 3 0,1564
0 3 0,0423
0 3 -0,1531
0 3 0,0591
0 4 -0,0647
0 3 0,1547
0 3 0,2363
0 3 0,1853
0 1 0,3674
0 3 0,1570
0 3 0,1148
0 4 -0,5099
0 3 0,1860
0 3 0,2129
0 3 0,1152
0 3 -0,0295
0 3 0,0909
0 3 -0,0568
0 5 -0,6613
0 3 -0,0052
0 3 0,1245
0 3 -0,1064
0 3 0,1390
0 3 0,1102
0 3 0,2678
0 3 0,0434
0 3 0,0237
0 3 0,1476
0 4 -0,4466
0 3 -0,0189
0 3 0,1378
0 3 0,0073
0 3 0,0708
0 3 -0,0574
0 3 0,2082
0 3 0,1802
0 3 0,0670
0 3 -0,1181
0 3 0,3001
0 3 -0,0095
0 3 0,0094
0 3 0,1734
0 4 -0,3211
0 4 -0,1145
0 3 0,1173
0 3 0,1310
0 3 0,2227
0 4 -0,1803
0 1 0,3138
0 3 0,2001
0 3 0,2337
0 1 0,4059
0 1 0,2000
0 4 -0,1756
0 1 0,2392
0 1 0,5176
0 3 0,2779
0 4 -0,2353
0 3 0,0053
0 3 0,2121
0 3 0,0060
0 3 0,2347
0 3 0,3463
0 1 0,3587
0 1 0,2988
0 1 0,3923
0 3 0,0350
0 1 0,3548
0 3 -0,0002
0 1 0,3481
0 3 0,1044
0 1 0,3269
0 1 0,2309
0 3 0,0204
0 3 0,2542
0 3 0,0353
0 1 0,3479
0 3 0,2073
0 1 0,4199
0 1 0,4902
0 3 0,1626
0 4 -0,3657
0 4 -0,2240
0 3 0,0862
0 3 0,1082
0 1 0,3621
0 4 -0,0487
0 1 0,3584
0 3 0,0234
0 3 0,1756
0 3 -0,0090
0 4 -0,1754
0 3 0,2881
0 3 0,1283
0 1 0,3026
0 4 -0,2721
0 3 -0,0486
0 3 0,0819
0 1 0,3632
0 3 0,0436
0 1 0,3625
0 3 0,1587
0 3 0,2143
0 3 0,1180
0 1 0,3601
0 3 0,1000
0 3 0,0222
0 1 0,3304
0 1 0,2650
0 4 -0,0365
0 1 0,4643
0 3 0,1424
0 1 0,2916
0 3 0,1652
0 3 0,1208
0 4 -0,2749
0 1 0,3942
0 4 -0,3477
0 3 0,2235
0 3 -0,0276
0 3 0,1783
0 3 0,2014
0 3 0,1829
0 3 0,0685
0 3 0,1516
0 1 0,4889
0 3 -0,0419
0 3 -0,0580
0 3 0,2257
0 1 0,3520
0 1 0,3327
0 3 0,1326
0 3 -0,1080
0 1 0,3992
0 1 0,4039
0 1 0,3585
0 3 0,0347
0 3 0,2124
0 3 0,0782
0 1 0,4087
0 4 -0,1200
0 3 0,0214
0 4 -0,2910
0 3 -0,0889
0 3 0,0597
0 3 0,2454
0 1 0,4189
0 3 0,0675
0 1 0,5479
0 3 0,0540
1 1 0,4101
1 4 -0,1791
1 3 0,0133
1 3 -0,1032
1 3 -0,0284
1 3 0,1193
1 1 0,3862
1 3 0,1683
1 3 0,1450
1 3 0,3306
1 3 0,3422
1 1 0,6084
1 1 0,5408
1 4 -0,1507
1 3 0,1205
1 3 0,1898
1 3 0,2660
1 1 0,4668
1 1 0,5824
1 4 -0,1792
1 4 -0,1503
1 3 0,2211
1 1 0,5183
1 3 0,0959
1 1 0,3350
1 4 -0,4501
1 3 0,1150
1 1 0,2804
1 3 0,0184
1 1 0,3154
1 1 0,3345
1 1 0,4233
1 3 0,0362
1 4 -0,0609
1 1 0,4282
1 3 0,1513
0 3 0,3447
0 3 -0,1513
0 1 0,2229
0 3 -0,0272
0 1 0,4681
0 3 0,0662
0 1 0,3277
0 3 0,3271
0 3 -0,0282
0 1 0,2924
0 3 0,0219
0 1 0,3364
0 3 0,0120
0 3 0,0376
0 3 0,0450
0 1 0,2930
0 3 0,2060
0 3 0,1596
0 3 0,2010
0 3 0,1567
0 3 0,2815
0 3 0,0829
0 4 -0,1490
0 3 0,0877
0 4 -0,0842
0 3 -0,0752
0 3 0,2273
0 1 0,4326
0 1 0,5278
0 3 0,0072
0 1 0,3470
0 3 0,1226
0 1 0,3753
0 1 0,4373
0 4 -0,1419
0 3 0,3240
0 3 0,0127
0 3 0,3439
0 3 -0,0262
0 3 -0,0343
0 1 0,3682
0 3 0,2696
0 3 0,0289
0 1 0,4115
0 3 0,0001
0 4 -0,1164
0 3 0,2955
0 3 -0,0107
0 3 0,2078
0 1 0,4393
0 5 -0,6130
0 3 0,1795
0 1 0,4264
0 3 0,0988
0 1 0,4169
0 1 0,2991
0 3 0,2579
0 3 0,2231
0 1 0,3771
0 3 0,2890
0 3 0,2319
0 3 0,1552
0 3 -0,0058
0 3 0,2596
0 3 0,1255
0 3 0,2194
0 3 0,2841
0 4 -0,1890
0 3 -0,1193
0 4 -0,1608
0 4 -0,2750
0 4 -0,1331
0 3 0,1066
0 1 0,3274
0 4 -0,4451
0 1 0,3537
0 4 -0,2848
0 3 0,0727
0 3 0,2157
0 3 0,2290
0 3 0,0080
0 3 0,0488
0 1 0,3031
0 3 0,0007
0 3 0,0374
0 3 -0,0780
0 3 0,0405
0 3 0,0629
0 3 -0,0581
0 3 0,2767
0 3 0,1615
0 4 -0,2402
0 3 0,1271
0 3 0,0863
0 1 0,3365
0 3 0,2287
0 4 -0,2188
0 1 0,2613
0 4 -0,1339
0 3 0,2490
0 3 0,2572
0 3 0,2223
0 4 -0,2619
0 1 0,4511
0 4 -0,1857
0 3 -0,0500
0 4 -0,1631
0 3 0,2613
0 1 0,3829
0 4 -0,3210
0 1 0,4862
0 4 -0,1515
0 1 0,2038
0 1 0,2967
0 3 -0,0169
0 3 0,1146
0 4 -0,3070
0 3 0,3024
0 4 -0,2233
0 4 -0,1848
0 4 -0,0246
0 4 -0,3028
0 3 0,1099
0 4 -0,0851
0 4 -0,1230
0 4 -0,2838
0 4 -0,1192
0 4 -0,0554
0 4 -0,0482
0 4 -0,2518
0 4 -0,3605
0 4 -0,2353
0 3 0,1667
0 3 -0,0208
0 4 -0,3490
0 3 0,1059
0 1 0,3054
0 3 0,2291
0 1 0,3317
0 4 -0,3669
0 1 0,5179
0 3 0,2673
0 3 0,1710
0 1 0,4133
0 1 0,3974
0 1 0,3871
0 1 0,2809
0 4 -0,1750
0 3 -0,0785
0 3 0,0663
0 3 0,2044
0 1 0,3968
0 3 0,0123
0 1 0,3553
0 4 -0,2643
0 3 0,1750
0 4 -0,4532
0 3 0,1487
0 3 0,2585
0 3 0,1618
0 1 0,4967
0 3 -0,0329
0 1 0,3088
0 3 0,2935
0 3 0,1601
0 3 -0,0020
0 3 -0,0231
0 3 0,1509
0 4 -0,1249
0 3 0,0689
0 3 -0,0736
0 3 -0,0335
0 4 -0,3107
0 3 0,2792
0 1 0,2517
0 3 0,1879
0 1 0,2742
0 4 -0,0843
0 1 0,3730
0 4 -0,0930
0 3 0,1502
0 3 0,1728
0 1 0,3693
0 1 0,3009
0 1 0,3140
0 1 0,3421
0 4 -0,1842
0 3 0,1470
0 4 -0,1555
0 1 0,4350
0 3 0,1321
0 3 0,0825
0 3 0,1511
0 1 0,3059
0 3 0,0867
0 3 0,2568
0 3 0,2271
0 3 0,0712
0 3 0,3033
0 3 0,2870
0 3 0,2011
0 1 0,1716
0 4 -0,1542
0 3 0,1218
0 3 0,1877
0 3 0,0950
0 3 0,1023
0 3 0,2408
0 3 0,1322
0 4 0,0935
0 4 -0,0893
0 1 0,3751
0 3 0,1606
0 3 0,2709
0 3 0,0943
0 3 -0,0006
0 1 0,3781
0 4 -0,0461
0 4 -0,2448
0 3 -0,0195
0 3 0,2013
0 3 0,0288
0 3 0,1125
0 3 0,2142
0 3 0,3035
0 1 0,3633
0 5 -0,6245
0 1 0,3117
0 1 0,5569
0 4 -0,2263
0 1 0,4842
0 3 -0,0085
0 3 0,2322
0 3 0,0649
0 3 0,2457
0 3 0,0666
0 3 0,0836
0 3 0,2523
0 4 -0,4137
0 1 0,3109
0 4 -0,0620
0 3 0,1975
0 1 0,4986
0 3 0,0809
0 3 0,2516
0 1 0,3218
0 1 0,3901
0 1 0,3123
0 3 0,0788
0 3 0,2637
0 3 0,2470
0 3 0,2143
0 3 0,0478
0 4 -0,1175
0 4 -0,1230
0 1 0,1658
0 3 0,1897
0 3 0,2427
0 1 0,3355
0 3 0,2917
0 3 0,0093
0 4 -0,0355
0 3 0,1423
0 1 0,1789
0 1 0,3768
0 1 0,3074
1 1 0,4809
1 3 0,2316
1 1 0,3460
1 3 0,2767
1 3 0,0507
1 3 0,0542
1 1 0,4169
1 3 -0,0328
1 3 0,3479
1 3 0,2540
1 1 0,3493
1 1 0,4385
1 1 0,3917
1 3 -0,0624
1 3 0,0405
1 4 -0,2716
1 4 -0,3169
1 3 -0,0446
1 3 0,1471
1 3 0,1083
1 3 0,3247
1 1 0,4036
1 3 -0,0365
1 3 0,2087
1 3 -0,0917
1 3 0,2304
1 3 -0,0241
1 4 -0,3339
1 3 0,0197
1 3 0,0633
1 4 -0,0762
1 3 0,1211
1 3 0,3122
1 4 -0,1325
1 3 0,0684
1 3 0,2686
1 4 -0,0900
1 3 0,0628
1 3 0,2071 |