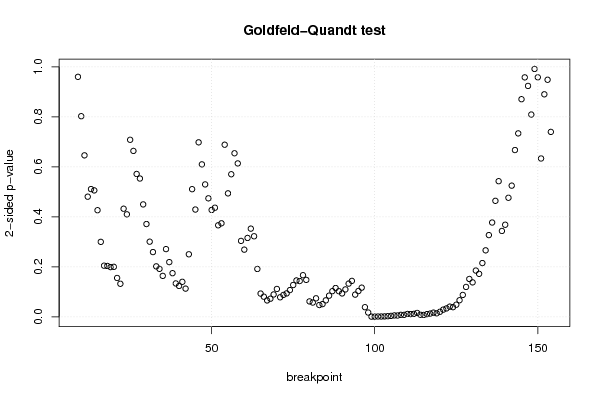
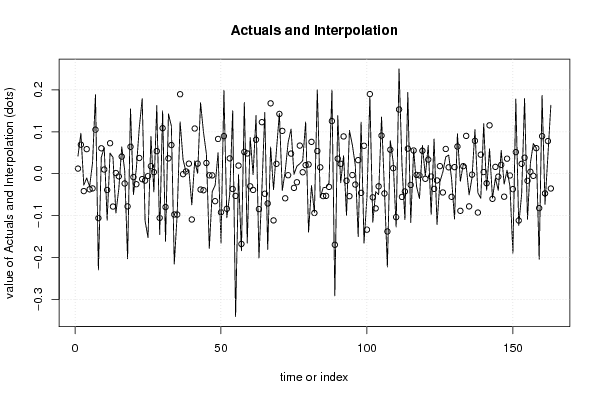
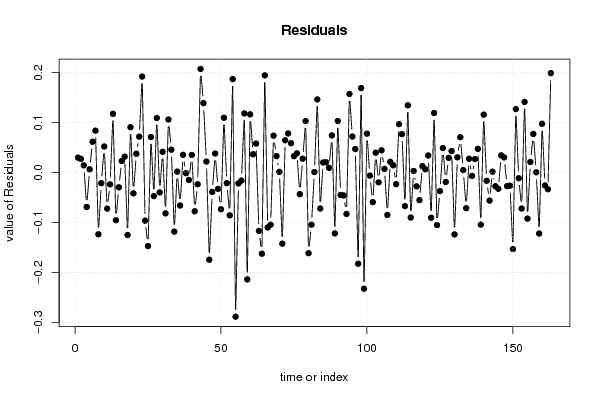
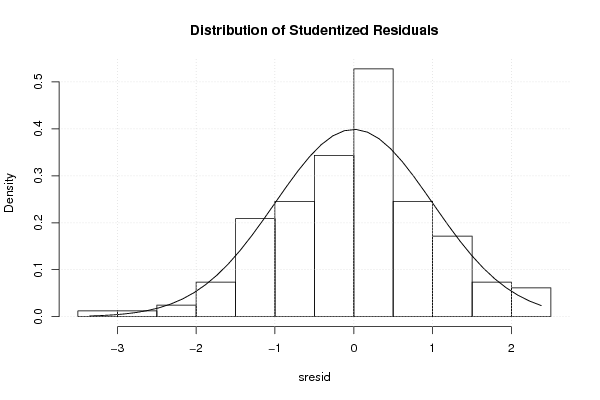
3,95508 78,70 56,90
4,15732 88,60 69,60
4,25277 104,20 82,60
4,16356 88,20 71,20
4,34640 94,70 74,10
4,27528 112,00 67,60
3,83514 78,90 56,30
4,11904 111,40 54,40
4,29456 132,50 65,00
4,31749 121,60 68,60
4,30946 116,10 80,30
4,31348 123,30 72,50
4,27249 107,90 88,30
4,19870 107,00 89,80
4,31882 115,80 103,50
4,21213 91,80 78,80
4,31214 93,50 85,70
4,33598 107,10 96,90
3,98713 80,50 66,90
4,24992 100,50 71,50
4,33205 100,20 89,50
4,37450 100,30 86,60
4,31482 96,60 91,20
4,17899 86,00 75,60
4,15104 76,90 75,60
4,16511 79,70 80,40
4,25277 93,10 91,60
4,31080 79,50 90,10
4,23989 80,30 90,80
4,31080 88,80 94,60
3,96651 72,40 62,60
4,11904 75,50 65,00
4,30271 92,90 87,60
4,37450 101,50 99,90
4,24563 94,70 85,10
4,34899 93,00 71,00
4,23989 79,80 73,00
4,31749 82,20 83,50
4,33598 87,60 86,50
4,32942 83,20 80,90
4,25277 81,60 80,80
4,49424 85,90 81,20
4,13517 71,90 61,90
4,07754 71,80 49,40
4,49424 98,30 79,20
4,42485 93,60 76,80
4,33073 86,10 81,20
4,45202 96,20 79,40
4,20320 78,60 74,00
4,32281 82,10 78,20
4,43793 94,40 98,90
4,40428 86,40 88,30
4,31749 82,20 79,30
4,52829 96,70 104,00
4,19570 84,20 60,50
4,32678 73,60 75,30
4,51415 94,90 106,20
4,48413 96,90 106,70
4,45202 90,20 95,40
4,46245 104,20 90,50
4,26268 78,40 113,80
4,42125 81,50 94,10
4,44265 96,70 109,90
4,37324 87,50 104,30
4,35028 86,20 80,70
4,56954 105,10 121,10
4,03424 72,90 68,80
4,32015 76,40 73,70
4,45783 100,50 104,20
4,44030 92,40 87,20
4,51743 96,30 94,50
4,70682 103,60 120,90
4,39445 75,10 88,50
4,40060 78,80 102,50
4,51086 93,70 118,60
4,39815 82,50 86,00
4,53796 88,30 110,60
4,61215 95,70 114,00
4,22683 73,30 72,60
4,35157 72,40 76,00
4,63181 94,00 114,60
4,72827 96,90 113,50
4,59006 92,40 115,20
4,68398 90,90 102,00
4,49536 93,50 101,50
4,53582 92,00 99,60
4,65014 115,90 113,80
4,54648 97,80 94,80
4,61215 97,70 102,00
4,71671 116,90 119,50
4,33205 96,70 88,00
4,62595 97,70 82,80
5,00529 103,90 112,10
5,14924 124,10 131,50
4,83310 117,30 110,00
4,88432 113,80 96,50
4,66814 100,00 101,90
4,75875 114,20 103,10
4,70773 116,30 103,50
4,80320 111,40 111,80
4,76388 103,40 100,30
4,81947 125,30 111,00
4,58497 92,50 84,60
4,53796 92,00 73,30
4,91486 121,60 112,00
4,87520 113,30 111,20
4,72916 92,50 82,40
4,61512 100,30 75,60
4,48526 83,20 64,20
4,57368 81,20 72,20
4,66155 94,50 80,80
4,55598 87,70 71,10
4,47734 82,30 153,20
4,67935 99,00 89,80
4,26409 72,40 57,30
4,28082 80,80 83,60
4,62006 105,50 88,40
4,63667 98,40 84,10
4,63473 94,50 95,50
4,48074 109,20 74,60
4,35671 84,10 79,80
4,51961 88,40 85,40
4,71402 111,30 106,40
4,60717 93,20 94,60
4,54648 86,30 94,60
4,77238 111,40 113,70
4,38826 85,40 66,70
4,52829 89,70 78,90
4,72827 110,90 126,30
4,71671 119,40 118,10
4,62203 109,30 117,30
4,66814 110,70 118,10
4,48751 101,30 108,60
4,61710 99,00 118,10
4,77912 117,90 141,00
4,65014 89,30 112,70
4,78916 105,40 131,90
4,72384 99,90 123,50
4,47847 79,50 81,30
4,59714 88,30 85,40
4,84024 116,20 138,50
4,72916 110,60 124,60
4,73795 99,30 125,80
4,85281 105,40 125,30
4,69318 89,90 111,00
4,67283 100,70 120,40
4,95794 122,50 141,40
4,66344 97,40 113,10
4,74667 97,90 114,00
4,86522 124,30 131,30
4,50424 94,70 77,80
4,57985 85,20 105,10
4,77322 101,90 125,40
4,79744 110,90 123,60
4,76644 102,00 107,90
4,65871 95,80 86,10
4,57780 86,90 97,80
4,58497 90,30 98,40
4,74319 97,90 118,00
4,69866 91,90 115,60
4,80320 90,40 114,50
4,81218 98,90 124,00
4,64535 81,30 101,80
4,60417 79,80 80,60
4,85593 93,70 129,70
4,84968 101,50 137,00
4,75961 88,60 127,30
4,71939 94,60 110,30
4,63279 84,20 134,90
4,70773 86,50 126,20
4,76899 92,60 130,50
4,80729 84,20 127,60
4,79082 85,90 134,80
4,78080 90,00 128,90
4,61809 79,10 101,10
4,61710 75,60 86,00
4,91339 97,00 139,20
4,89485 96,40 126,80
4,69684 85,20 117,10
4,75186 100,30 103,00
4,64727 76,70 108,70
4,74493 79,00 115,00
4,82511 94,40 133,20
4,81300 82,80 131,30
4,78749 74,60 119,60
4,88280 92,80 146,70
4,67470 69,70 101,00
4,61512 68,90 88,70
5,03109 97,50 143,70
4,97328 92,90 138,10
4,83469 93,40 139,80
4,83151 92,10 121,60
4,71582 80,60 112,60
4,77407 86,00 136,70
4,90971 93,60 147,40
4,87290 90,30 128,10
4,85593 81,30 117,50
4,92071 98,40 148,20
4,52287 73,30 101,60
4,64150 77,10 90,40
4,93447 91,40 148,60
4,82831 89,00 133,80
4,86907 94,10 130,30
4,75703 94,70 113,60
4,66721 80,70 105,80
4,79744 85,20 136,10
4,99451 107,90 160,30
4,75359 81,60 127,70
4,92362 83,80 141,80
4,91559 98,80 149,30
4,56226 75,60 94,50
4,84419 80,70 95,20
|