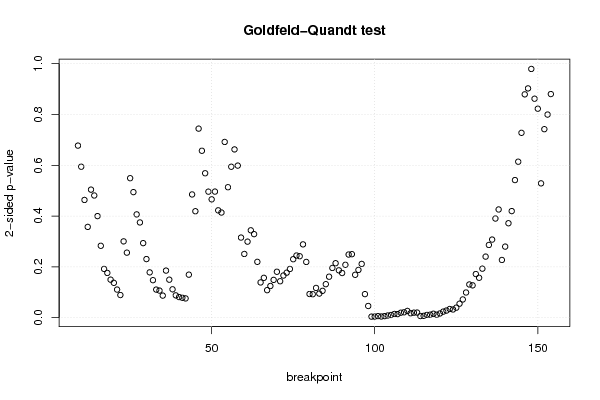
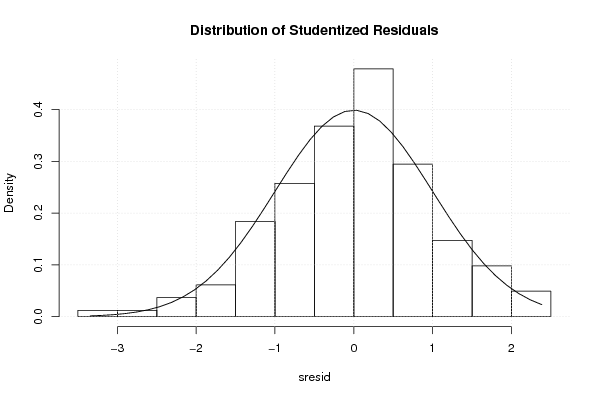
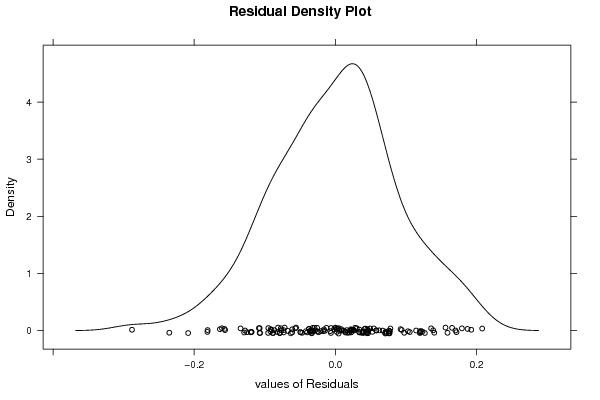
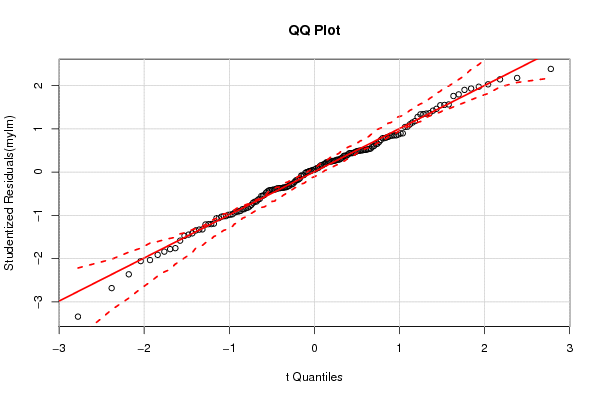
3,95508 0,02526 0,80199
4,15732 0,02286 0,79322
4,25277 0,01994 0,78584
4,16356 0,02295 0,79224
4,34640 0,02161 0,79051
4,27528 0,01876 0,79448
3,83514 0,02521 0,80246
4,11904 0,01885 0,80396
4,29456 0,01629 0,79619
4,31749 0,01751 0,79385
4,30946 0,01820 0,78705
4,31348 0,01730 0,79145
4,27249 0,01936 0,78298
4,19870 0,01950 0,78226
4,31882 0,01824 0,77622
4,21213 0,02219 0,78786
4,31214 0,02185 0,78426
4,33598 0,01948 0,77902
3,98713 0,02478 0,79494
4,24992 0,02056 0,79205
4,33205 0,02061 0,78240
4,37450 0,02059 0,78381
4,31482 0,02125 0,78160
4,17899 0,02344 0,78965
4,15104 0,02576 0,78965
4,16511 0,02499 0,78700
4,25277 0,02193 0,78141
4,31080 0,02505 0,78212
4,23989 0,02484 0,78179
4,31080 0,02282 0,78004
3,96651 0,02710 0,79782
4,11904 0,02616 0,79619
4,30271 0,02197 0,78332
4,37450 0,02039 0,77772
4,24563 0,02161 0,78456
4,34899 0,02195 0,79236
4,23989 0,02497 0,79116
4,31749 0,02435 0,78537
4,33598 0,02308 0,78386
4,32942 0,02410 0,78673
4,25277 0,02450 0,78678
4,49424 0,02346 0,78657
4,13517 0,02726 0,79831
4,07754 0,02729 0,80821
4,49424 0,02094 0,78764
4,42485 0,02183 0,78897
4,33073 0,02342 0,78657
4,45202 0,02133 0,78753
4,20320 0,02529 0,79057
4,32281 0,02438 0,78819
4,43793 0,02167 0,77815
4,40428 0,02335 0,78298
4,31749 0,02435 0,78759
4,52829 0,02124 0,77601
4,19570 0,02386 0,79931
4,32678 0,02673 0,78982
4,51415 0,02158 0,77513
4,48413 0,02120 0,77493
4,45202 0,02252 0,77968
4,46245 0,01994 0,78193
4,26268 0,02534 0,77221
4,42125 0,02453 0,78026
4,44265 0,02124 0,77368
4,37324 0,02310 0,77589
4,35028 0,02340 0,78684
4,56954 0,01980 0,76959
4,03424 0,02694 0,79372
4,32015 0,02590 0,79074
4,45783 0,02056 0,77593
4,44030 0,02207 0,78352
4,51743 0,02131 0,78008
4,70682 0,02004 0,76966
4,39445 0,02628 0,78288
4,40060 0,02523 0,77663
4,51086 0,02181 0,77047
4,39815 0,02428 0,78411
4,53796 0,02293 0,77341
4,61215 0,02142 0,77213
4,22683 0,02682 0,79139
4,35157 0,02710 0,78942
4,63181 0,02175 0,77191
4,72827 0,02120 0,77232
4,59006 0,02207 0,77169
4,68398 0,02237 0,77684
4,49536 0,02185 0,77705
4,53582 0,02215 0,77785
4,65014 0,01823 0,77221
4,54648 0,02103 0,77995
4,61215 0,02105 0,77684
4,71671 0,01810 0,77015
4,33205 0,02124 0,78313
4,62595 0,02105 0,78573
5,00529 0,01999 0,77284
5,14924 0,01721 0,76614
4,83310 0,01805 0,77364
4,88432 0,01851 0,77919
4,66814 0,02064 0,77688
4,75875 0,01846 0,77638
4,70773 0,01818 0,77622
4,80320 0,01885 0,77296
4,76388 0,02007 0,77755
4,81947 0,01707 0,77326
4,58497 0,02205 0,78481
4,53796 0,02215 0,79098
4,91486 0,01751 0,77288
4,87520 0,01858 0,77318
4,72916 0,02205 0,78594
4,61512 0,02059 0,78965
4,48526 0,02410 0,79673
4,57368 0,02460 0,79163
4,66155 0,02165 0,78678
4,55598 0,02306 0,79230
4,47734 0,02433 0,75977
4,67935 0,02082 0,78226
4,26409 0,02710 0,80169
4,28082 0,02471 0,78532
4,62006 0,01973 0,78293
4,63667 0,02093 0,78507
4,63473 0,02165 0,77964
4,48074 0,01917 0,79022
4,35671 0,02389 0,78732
4,51961 0,02290 0,78441
4,71402 0,01886 0,77505
4,60717 0,02191 0,78004
4,54648 0,02337 0,78004
4,77238 0,01885 0,77225
4,38826 0,02358 0,79507
4,52829 0,02262 0,78781
4,72827 0,01892 0,76783
4,71671 0,01778 0,77065
4,62203 0,01915 0,77093
4,66814 0,01895 0,77065
4,48751 0,02042 0,77418
4,61710 0,02082 0,77065
4,77912 0,01797 0,76323
4,65014 0,02271 0,77262
4,78916 0,01975 0,76601
4,72384 0,02066 0,76877
4,47847 0,02505 0,78652
4,59714 0,02293 0,78441
4,84024 0,01819 0,76397
4,72916 0,01896 0,76840
4,73795 0,02077 0,76799
4,85281 0,01975 0,76816
4,69318 0,02258 0,77326
4,67283 0,02052 0,76984
4,95794 0,01740 0,76311
4,66344 0,02111 0,77247
4,74667 0,02102 0,77213
4,86522 0,01719 0,76620
4,50424 0,02161 0,78841
4,57985 0,02363 0,77557
4,77322 0,02032 0,76813
4,79744 0,01892 0,76873
4,76644 0,02030 0,77446
4,65871 0,02140 0,78406
4,57780 0,02324 0,77862
4,58497 0,02250 0,77836
4,74319 0,02102 0,77068
4,69866 0,02217 0,77155
4,80320 0,02248 0,77195
4,81218 0,02084 0,76860
4,64535 0,02458 0,77692
4,60417 0,02497 0,78689
4,85593 0,02181 0,76671
4,84968 0,02039 0,76443
4,75961 0,02286 0,76750
4,71939 0,02163 0,77353
4,63279 0,02386 0,76507
4,70773 0,02333 0,76786
4,76899 0,02203 0,76646
4,80729 0,02386 0,76740
4,79082 0,02346 0,76510
4,78080 0,02256 0,76697
4,61809 0,02515 0,77721
4,61710 0,02613 0,78411
4,91339 0,02118 0,76376
4,89485 0,02129 0,76766
4,69684 0,02363 0,77100
4,75186 0,02059 0,77642
4,64727 0,02581 0,77414
4,74493 0,02518 0,77177
4,82511 0,02167 0,76560
4,81300 0,02420 0,76620
4,78749 0,02643 0,77012
4,88280 0,02199 0,76158
4,67470 0,02798 0,77726
4,61512 0,02826 0,78279
5,03109 0,02109 0,76244
4,97328 0,02197 0,76409
4,83469 0,02187 0,76358
4,83151 0,02213 0,76942
4,71582 0,02476 0,77266
4,77407 0,02344 0,76452
4,90971 0,02183 0,76138
4,87290 0,02250 0,76723
4,85593 0,02458 0,77086
4,92071 0,02093 0,76115
4,52287 0,02682 0,77700
4,64150 0,02570 0,78198
4,93447 0,02227 0,76104
4,82831 0,02277 0,76541
4,86907 0,02173 0,76652
4,75703 0,02161 0,77228
4,66721 0,02473 0,77529
4,79744 0,02363 0,76470
4,99451 0,01936 0,75790
4,75359 0,02450 0,76736
4,92362 0,02396 0,76299
4,91559 0,02086 0,76085
4,56226 0,02613 0,78008
4,84419 0,02473 0,77977
|