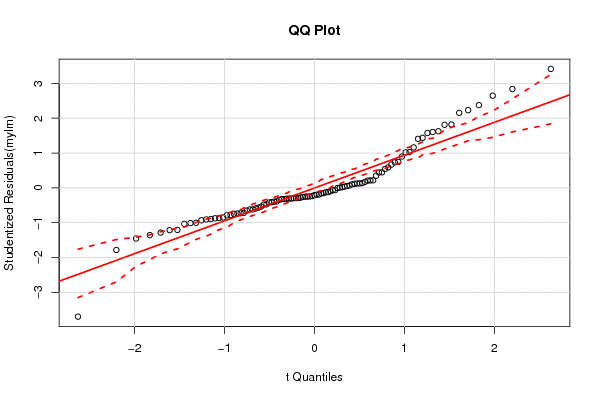
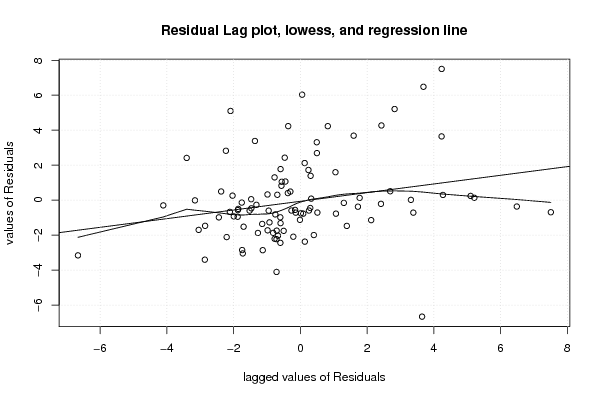
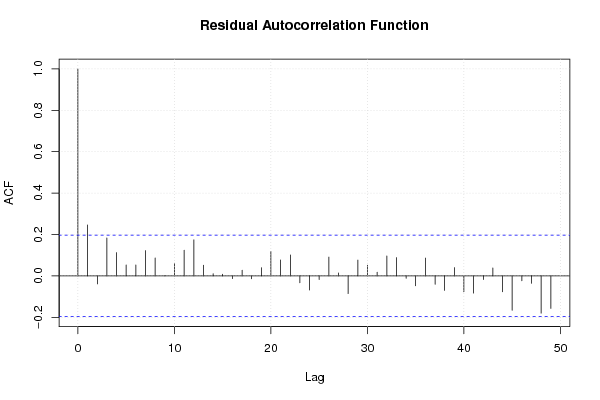
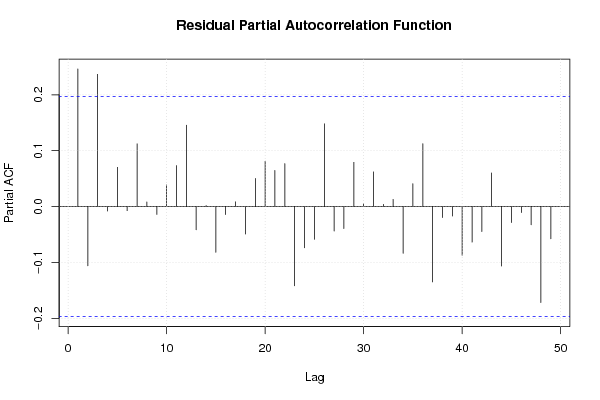
1 0,455 0,365 0,095 0,514 0,2245 0,101 0,15 15
1 0,35 0,265 0,09 0,2255 0,0995 0,0485 0,07 7
-1 0,53 0,42 0,135 0,677 0,2565 0,1415 0,21 9
1 0,44 0,365 0,125 0,516 0,2155 0,114 0,155 10
0 0,33 0,255 0,08 0,205 0,0895 0,0395 0,055 7
0 0,425 0,3 0,095 0,3515 0,141 0,0775 0,12 8
-1 0,53 0,415 0,15 0,7775 0,237 0,1415 0,33 20
-1 0,545 0,425 0,125 0,768 0,294 0,1495 0,26 16
1 0,475 0,37 0,125 0,5095 0,2165 0,1125 0,165 9
-1 0,55 0,44 0,15 0,8945 0,3145 0,151 0,32 19
-1 0,525 0,38 0,14 0,6065 0,194 0,1475 0,21 14
1 0,43 0,35 0,11 0,406 0,1675 0,081 0,135 10
1 0,49 0,38 0,135 0,5415 0,2175 0,095 0,19 11
-1 0,535 0,405 0,145 0,6845 0,2725 0,171 0,205 10
-1 0,47 0,355 0,1 0,4755 0,1675 0,0805 0,185 10
1 0,5 0,4 0,13 0,6645 0,258 0,133 0,24 12
0 0,355 0,28 0,085 0,2905 0,095 0,0395 0,115 7
-1 0,44 0,34 0,1 0,451 0,188 0,087 0,13 10
1 0,365 0,295 0,08 0,2555 0,097 0,043 0,1 7
1 0,45 0,32 0,1 0,381 0,1705 0,075 0,115 9
1 0,355 0,28 0,095 0,2455 0,0955 0,062 0,075 11
0 0,38 0,275 0,1 0,2255 0,08 0,049 0,085 10
-1 0,565 0,44 0,155 0,9395 0,4275 0,214 0,27 12
-1 0,55 0,415 0,135 0,7635 0,318 0,21 0,2 9
-1 0,615 0,48 0,165 1,1615 0,513 0,301 0,305 10
-1 0,56 0,44 0,14 0,9285 0,3825 0,188 0,3 11
-1 0,58 0,45 0,185 0,9955 0,3945 0,272 0,285 11
1 0,59 0,445 0,14 0,931 0,356 0,234 0,28 12
1 0,605 0,475 0,18 0,9365 0,394 0,219 0,295 15
1 0,575 0,425 0,14 0,8635 0,393 0,227 0,2 11
1 0,58 0,47 0,165 0,9975 0,3935 0,242 0,33 10
-1 0,68 0,56 0,165 1,639 0,6055 0,2805 0,46 15
1 0,665 0,525 0,165 1,338 0,5515 0,3575 0,35 18
-1 0,68 0,55 0,175 1,798 0,815 0,3925 0,455 19
-1 0,705 0,55 0,2 1,7095 0,633 0,4115 0,49 13
1 0,465 0,355 0,105 0,4795 0,227 0,124 0,125 8
-1 0,54 0,475 0,155 1,217 0,5305 0,3075 0,34 16
-1 0,45 0,355 0,105 0,5225 0,237 0,1165 0,145 8
-1 0,575 0,445 0,135 0,883 0,381 0,2035 0,26 11
1 0,355 0,29 0,09 0,3275 0,134 0,086 0,09 9
-1 0,45 0,335 0,105 0,425 0,1865 0,091 0,115 9
-1 0,55 0,425 0,135 0,8515 0,362 0,196 0,27 14
0 0,24 0,175 0,045 0,07 0,0315 0,0235 0,02 5
0 0,205 0,15 0,055 0,042 0,0255 0,015 0,012 5
0 0,21 0,15 0,05 0,042 0,0175 0,0125 0,015 4
0 0,39 0,295 0,095 0,203 0,0875 0,045 0,075 7
1 0,47 0,37 0,12 0,5795 0,293 0,227 0,14 9
-1 0,46 0,375 0,12 0,4605 0,1775 0,11 0,15 7
0 0,325 0,245 0,07 0,161 0,0755 0,0255 0,045 6
-1 0,525 0,425 0,16 0,8355 0,3545 0,2135 0,245 9
0 0,52 0,41 0,12 0,595 0,2385 0,111 0,19 8
1 0,4 0,32 0,095 0,303 0,1335 0,06 0,1 7
1 0,485 0,36 0,13 0,5415 0,2595 0,096 0,16 10
-1 0,47 0,36 0,12 0,4775 0,2105 0,1055 0,15 10
1 0,405 0,31 0,1 0,385 0,173 0,0915 0,11 7
-1 0,5 0,4 0,14 0,6615 0,2565 0,1755 0,22 8
1 0,445 0,35 0,12 0,4425 0,192 0,0955 0,135 8
1 0,47 0,385 0,135 0,5895 0,2765 0,12 0,17 8
0 0,245 0,19 0,06 0,086 0,042 0,014 0,025 4
-1 0,505 0,4 0,125 0,583 0,246 0,13 0,175 7
1 0,45 0,345 0,105 0,4115 0,18 0,1125 0,135 7
1 0,505 0,405 0,11 0,625 0,305 0,16 0,175 9
-1 0,53 0,41 0,13 0,6965 0,302 0,1935 0,2 10
1 0,425 0,325 0,095 0,3785 0,1705 0,08 0,1 7
1 0,52 0,4 0,12 0,58 0,234 0,1315 0,185 8
1 0,475 0,355 0,12 0,48 0,234 0,1015 0,135 8
-1 0,565 0,44 0,16 0,915 0,354 0,1935 0,32 12
-1 0,595 0,495 0,185 1,285 0,416 0,224 0,485 13
-1 0,475 0,39 0,12 0,5305 0,2135 0,1155 0,17 10
0 0,31 0,235 0,07 0,151 0,063 0,0405 0,045 6
1 0,555 0,425 0,13 0,7665 0,264 0,168 0,275 13
-1 0,4 0,32 0,11 0,353 0,1405 0,0985 0,1 8
-1 0,595 0,475 0,17 1,247 0,48 0,225 0,425 20
1 0,57 0,48 0,175 1,185 0,474 0,261 0,38 11
-1 0,605 0,45 0,195 1,098 0,481 0,2895 0,315 13
-1 0,6 0,475 0,15 1,0075 0,4425 0,221 0,28 15
1 0,595 0,475 0,14 0,944 0,3625 0,189 0,315 9
-1 0,6 0,47 0,15 0,922 0,363 0,194 0,305 10
-1 0,555 0,425 0,14 0,788 0,282 0,1595 0,285 11
-1 0,615 0,475 0,17 1,1025 0,4695 0,2355 0,345 14
-1 0,575 0,445 0,14 0,941 0,3845 0,252 0,285 9
1 0,62 0,51 0,175 1,615 0,5105 0,192 0,675 12
-1 0,52 0,425 0,165 0,9885 0,396 0,225 0,32 16
1 0,595 0,475 0,16 1,3175 0,408 0,234 0,58 21
1 0,58 0,45 0,14 1,013 0,38 0,216 0,36 14
-1 0,57 0,465 0,18 1,295 0,339 0,2225 0,44 12
1 0,625 0,465 0,14 1,195 0,4825 0,205 0,4 13
1 0,56 0,44 0,16 0,8645 0,3305 0,2075 0,26 10
-1 0,46 0,355 0,13 0,517 0,2205 0,114 0,165 9
-1 0,575 0,45 0,16 0,9775 0,3135 0,231 0,33 12
1 0,565 0,425 0,135 0,8115 0,341 0,1675 0,255 15
1 0,555 0,44 0,15 0,755 0,307 0,1525 0,26 12
1 0,595 0,465 0,175 1,115 0,4015 0,254 0,39 13
-1 0,625 0,495 0,165 1,262 0,507 0,318 0,39 10
1 0,695 0,56 0,19 1,494 0,588 0,3425 0,485 15
1 0,665 0,535 0,195 1,606 0,5755 0,388 0,48 14
1 0,535 0,435 0,15 0,725 0,269 0,1385 0,25 9
1 0,47 0,375 0,13 0,523 0,214 0,132 0,145 8
1 0,47 0,37 0,13 0,5225 0,201 0,133 0,165 7 |