Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_density.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
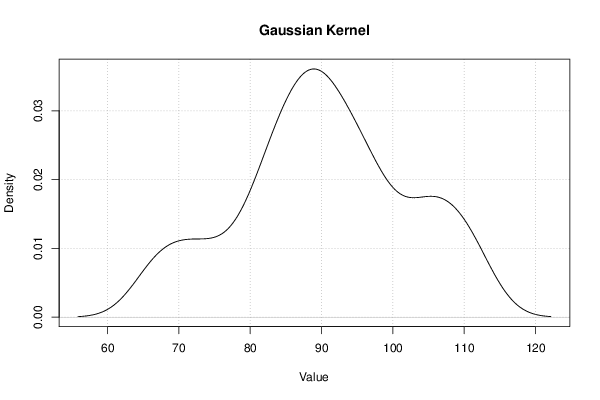
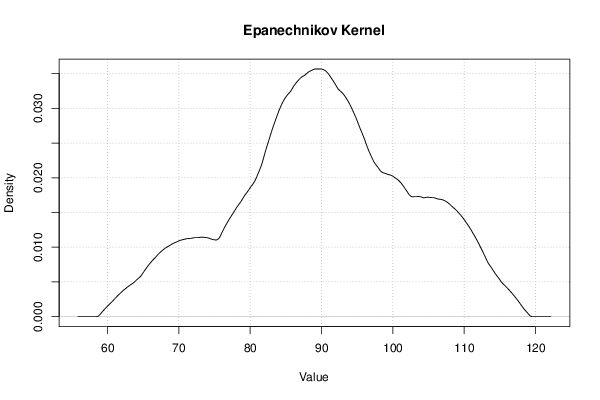
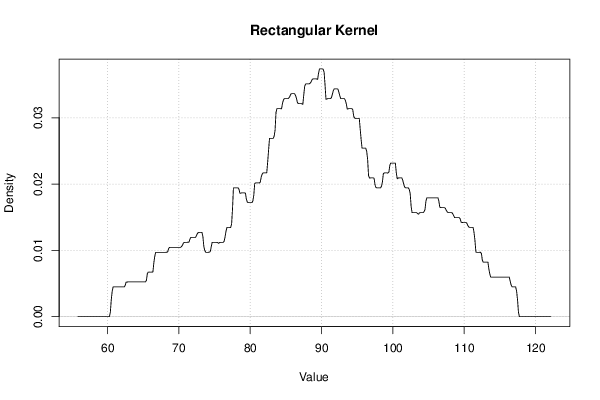
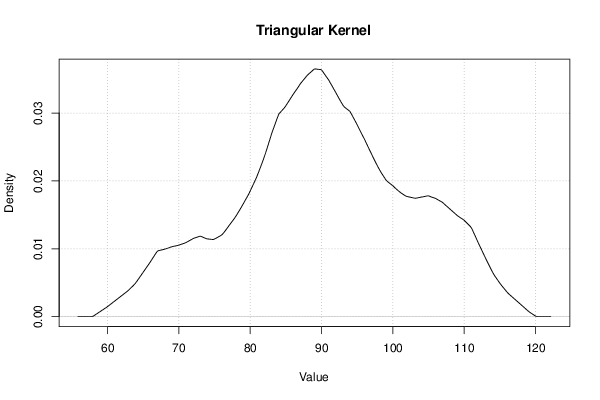
| Title produced by software | Kernel Density Estimation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 27 Oct 2014 14:15:17 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2014/Oct/27/t1414419336xzyex9nse1brjjt.htm/, Retrieved Fri, 10 May 2024 20:39:01 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=247044, Retrieved Fri, 10 May 2024 20:39:01 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 72 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Boxplot and Trimmed Means] [Care Age 10 Data] [2009-10-26 09:01:50] [98fd0e87c3eb04e0cc2efde01dbafab6] - PD [Boxplot and Trimmed Means] [Care Age 7 Data] [2009-10-26 18:36:29] [98fd0e87c3eb04e0cc2efde01dbafab6] - P [CARE Data - Boxplots and Scatterplot Matrix] [CARE Data] [2010-10-19 14:16:27] [3fdd735c61ad38cbc9b3393dc997cdb7] - RM [CARE Data - Boxplots and Scatterplot Matrix] [CARE data - works...] [2011-10-17 10:23:12] [98fd0e87c3eb04e0cc2efde01dbafab6] - RMP [Boxplot and Trimmed Means] [CARE Study Age 7 ] [2013-10-17 12:59:45] [34296d8f7657c52ed60d5bff9133afec] - RM [Boxplot and Trimmed Means] [year 7 data] [2014-10-23 10:48:16] [770cabeb704b463c26ebb6813270a402] - M D [Boxplot and Trimmed Means] [Year 10data] [2014-10-27 13:53:03] [770cabeb704b463c26ebb6813270a402] - D [Boxplot and Trimmed Means] [AFS] [2014-10-27 14:05:09] [770cabeb704b463c26ebb6813270a402] - RM [Kernel Density Estimation] [Maternal IQ] [2014-10-27 14:09:12] [770cabeb704b463c26ebb6813270a402] - R D [Kernel Density Estimation] [AVA] [2014-10-27 14:11:46] [770cabeb704b463c26ebb6813270a402] - D [Kernel Density Estimation] [ARD] [2014-10-27 14:14:01] [770cabeb704b463c26ebb6813270a402] - D [Kernel Density Estimation] [AMA] [2014-10-27 14:15:17] [f06adca6bfb99da24b496db9458d00d0] [Current] - D [Kernel Density Estimation] [AKN] [2014-10-27 14:16:29] [770cabeb704b463c26ebb6813270a402] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
81 90 91 87 89 85 81 80 75 84 91 87 90 72 96 88 73 86 67 89 94 88 77 89 104 89 107 96 111 84 83 82 105 100 111 90 87 90 67 84 84 96 67 90 83 78 100 67 94 89 89 111 88 94 111 111 105 97 83 105 69 80 87 67 92 102 90 98 94 101 96 94 105 105 84 72 96 94 107 103 95 73 110 84 67 73 84 79 91 92 107 111 89 110 98 95 84 73 86 98 90 97 106 106 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
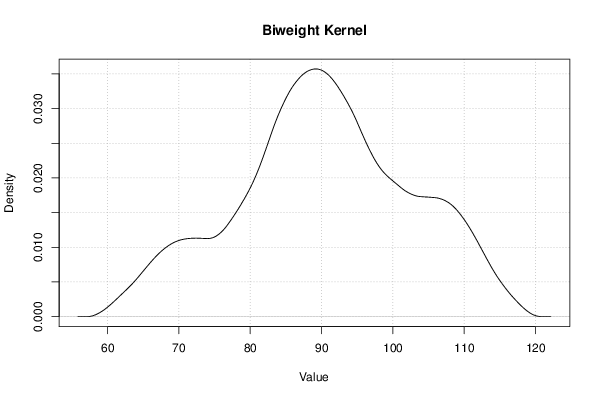
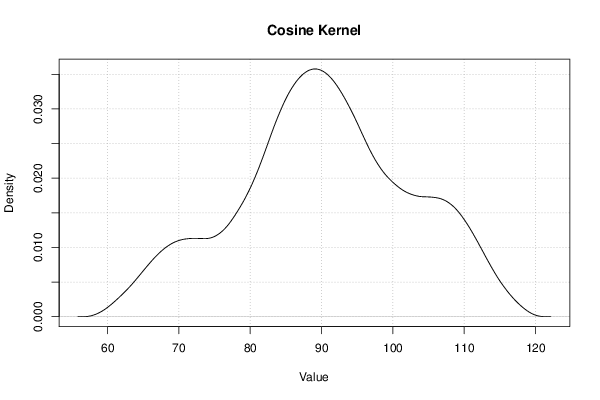
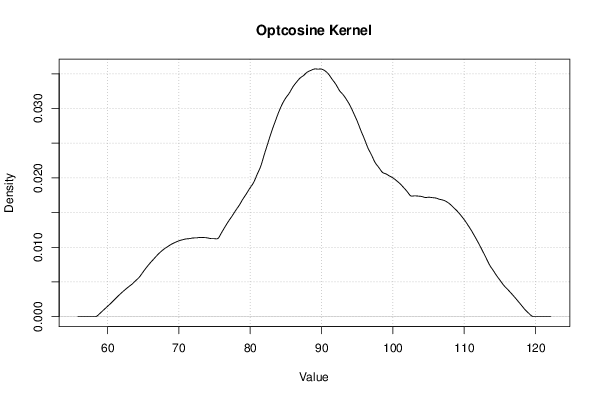
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 2 ; par2 = TRUE ; par3 = 5 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 2 ; par2 = TRUE ; par3 = 5 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par3 <- '512' | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||