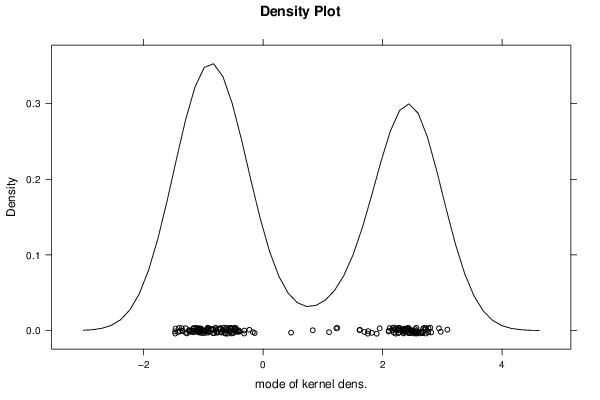
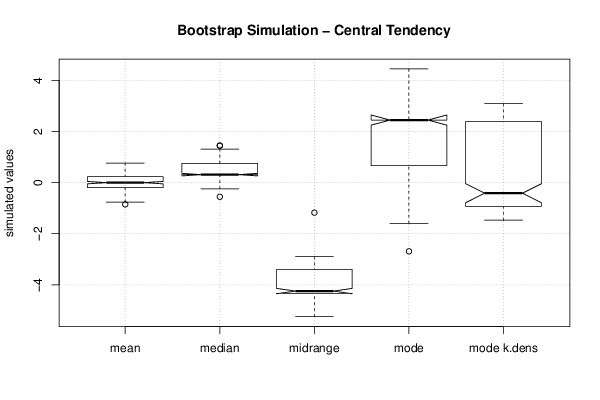
2,30928
1,44306
3,30928
-2,55694
2,44306
-8,55694
1,30928
1,44306
0,443064
-1,55694
1,44306
-5,55694
-0,556936
0,30928
-0,69072
-4,24631
-0,556936
2,30928
-0,11253
-3,69072
-4,55694
2,44306
2,30928
-2,55694
4,44306
-11,5569
8,88747
0,753686
2,44306
-2,69072
-2,69072
0,30928
4,44306
-2,55694
2,44306
0,443064
-8,69072
-7,11253
1,44306
7,30928
2,44306
2,44306
3,44306
3,44306
-3,11253
4,30928
-5,55694
-2,11253
-2,69072
3,88747
1,88747
-4,11253
-1,55694
-0,11253
-2,55694
6,44306
1,75369
-7,55694
-11,2463
7,88747
7,44306
4,30928
2,30928
-1,55694
0,30928
-1,11253
-0,69072
-1,55694
-3,55694
-0,246314
6,30928
0,30928
2,75369
1,88747
-4,69072
7,44306
-2,69072
2,88747
1,30928
-7,24631
3,44306
2,75369
-7,24631
2,75369
-1,11253
-9,24631
1,88747
-4,11253
2,75369
4,75369
-1,24631
-0,11253
-1,11253
-1,11253
2,75369
-2,11253
0,753686
4,88747
1,88747
2,88747
-0,11253
-0,11253
-0,246314
-13,1125
-4,11253
-1,11253
-8,24631
-11,2463
-4,24631
-1,24631
-1,24631
-0,246314
-2,11253
2,44306
1,44306
0,443064
4,44306
10,7537
-4,11253
6,44306
4,30928
2,44306
-0,69072
-0,69072
2,44306
-1,55694
-5,55694
-0,556936
-4,55694
2,88747
4,44306
4,44306
0,30928
-1,55694
-4,55694
-1,55694
-1,55694
2,44306
0,443064
3,30928
-1,55694
-1,11253
-0,556936
-17,5569
2,44306
4,30928
1,30928
-5,55694
-2,69072
-11,6907
3,44306
-1,69072
3,44306
3,44306
0,30928
3,88747
-1,11253
7,44306
4,30928
7,75369
-1,24631
8,75369
-0,11253
0,30928
2,44306
7,75369
0,88747
8,75369
3,88747
1,75369
2,75369
6,88747
0,753686
7,88747
4,44306
4,44306
-1,11253
-0,556936
-2,69072
-1,11253
1,30928
3,75369
-1,24631
4,44306
-1,24631
-2,11253
5,75369
5,75369
5,75369
4,75369
-2,11253
4,88747
-11,1125
-4,11253
-6,24631
-2,24631
8,88747
6,30928
-15,2463
-3,24631
1,75369
2,44306
0,88747
-1,69072
0,753686
-0,556936
2,75369
-4,55694
1,88747
-19,2463
-3,11253
3,88747
2,44306
-6,69072
-1,24631
5,30928
-9,55694
-3,11253
2,44306
2,88747
9,75369
-0,69072
-4,24631
8,44306
-4,55694
0,30928
2,75369
-2,69072
1,88747
2,44306
0,30928
-14,6907
1,30928
2,88747
-0,556936
-16,5569
3,44306
2,75369
-4,55694
-17,5569
-6,11253
5,30928
-2,24631
-1,11253
7,75369
4,88747
1,88747
-1,69072
-0,556936
3,30928
-1,55694
3,44306
0,30928
1,88747
-4,24631
6,44306
6,75369
0,88747
3,30928
-1,24631
-6,11253
0,88747
7,75369
-11,1125
-1,11253
-2,24631
1,88747
-0,246314
-6,24631
5,88747
1,88747
-3,24631
3,88747
-17,2463
6,75369
-1,11253
4,30928
2,75369
2,88747
|