Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_bootstrapplot1.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Bootstrap Plot - Central Tendency | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 15 Dec 2014 20:01:24 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2014/Dec/15/t1418673705e1dpzl7b0zdu39k.htm/, Retrieved Sun, 19 May 2024 13:34:45 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=268968, Retrieved Sun, 19 May 2024 13:34:45 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 82 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Central Tendency] [] [2014-11-04 10:08:09] [32b17a345b130fdf5cc88718ed94a974] - [Central Tendency] [WS7 3] [2014-11-12 20:26:33] [81f624c2f0b20a2549c93e7c3dccf981] - D [Central Tendency] [] [2014-12-15 16:40:00] [ae96d02647dd9ad9c105f1fa6642e295] - RM D [Bootstrap Plot - Central Tendency] [] [2014-12-15 20:01:24] [4ade5e15fc88dfcb6333f94ac70b9a75] [Current] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
0.0991579 -0.24856 -0.129539 -5.65751 -6.22954 -0.265554 1.6447 0.434446 -2.27816 -4.98621 -1.59352 -6.75458 -2.16921 -1.12149 -6.5023 -1.78987 -1.12515 -3.72588 -3.33027 -2.89718 -6.88182 0.74249 -2.41856 1.28818 -1.4553 -2.02954 3.04104 0.310134 -3.09352 -1.65385 -4.69425 -1.42222 -3.99352 -3.2928 -2.28987 -2.40595 -2.14563 -0.453851 -3.62954 -0.905954 -6.99718 -1.36921 -0.284001 -2.28987 -0.822221 -1.21327 -1.28987 -5.09352 0.25144 -0.565554 -1.32954 -6.26262 -2.18987 -1.24653 -0.0141772 -2.79718 -7.48766 1.56387 -4.57653 0.210134 -3.72588 -0.172872 -4.97726 2.81013 -3.66628 -3.6045 -1.79718 0.038105 1.63445 -0.793524 -0.886207 -1.49718 -3.93758 2.11981 0.150714 -0.0582362 -0.104501 0.17412 -3.09425 -5.38987 -2.42954 0.534446 -2.18987 -8.56555 -2.88693 -1.32222 -1.88987 -1.2736 -4.13686 0.495499 -0.298636 -7.20157 -3.38182 -4.1619 -5.1332 -4.08987 -5.69352 -1.3045 1.24851 -4.70889 -2.42222 -2.78987 -4.21182 -3.61784 1.36608 -4.65458 0.891114 -8.0619 -6.40157 -3.07287 -1.39352 -1.15515 -8.83693 -0.422221 5.01013 4.76013 3.73372 -0.393524 4.20282 6.10648 2.94542 0.646226 5.82347 1.61086 -2.45824 -0.58548 3.01307 0.221838 5.81013 0.946149 1.41013 3.15648 1.81973 1.61307 -0.542798 -0.439866 4.16379 -4.77816 5.47046 2.91379 -0.720589 4.72485 2.25648 4.56013 3.26013 4.72046 0.510134 1.26013 1.66013 5.35282 -3.12661 2.78071 5.16013 3.85648 1.47778 0.695423 5.6351 2.60648 1.79249 -1.33987 5.36013 2.64908 4.1381 2.7851 7.3551 -2.04352 5.45575 2.10648 2.10209 -1.77222 2.57184 5.07339 1.76608 2.37412 2.37412 4.57412 0.29249 6.33006 2.26013 -4.75222 0.662417 1.22568 6.07412 2.09542 -0.158963 3.10648 4.5168 0.25209 -0.619213 -0.619289 -2.67661 2.27778 -3.29718 5.14176 1.02347 1.76013 1.72339 0.977779 2.06608 3.09542 6.22485 0.509408 0.605749 2.49542 0.174196 1.60648 -3.40019 -0.280266 6.686 4.24778 -1.98621 1.9351 -1.19059 0.880864 3.54916 -0.400842 2.79176 4.89981 -2.03693 2.75282 4.72412 -5.66418 -0.772221 2.79843 6.21013 2.46242 4.1072 2.60648 5.08582 6.05648 5.55575 6.10575 -0.0538508 -0.20751 -3.87816 -8.62222 -0.727257 0.734446 -1.32588 -0.914177 0.324846 -1.75385 1.81013 7.3551 -1.3758 1.60575 4.22111 1.28151 3.17046 0.32412 -1.11117 -0.947183 1.66013 1.89908 0.398431 3.79176 -5.30385 -5.26117 -0.00888631 -5.27222 -2.04352 -0.387583 -3.13987 1.81013 3.56013 0.405749 1.05282 2.61013 3.72778 -2.33255 2.25868 -1.43701 2.04176 4.66013 2.04542 1.51013 3.84843 -4.51034 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
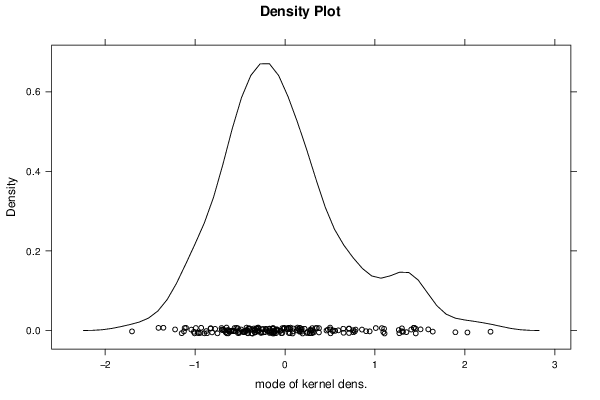
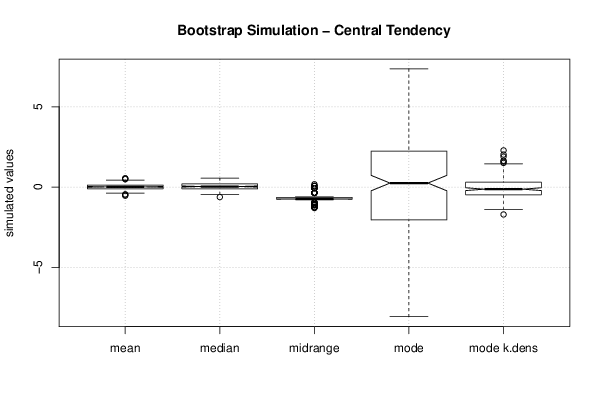
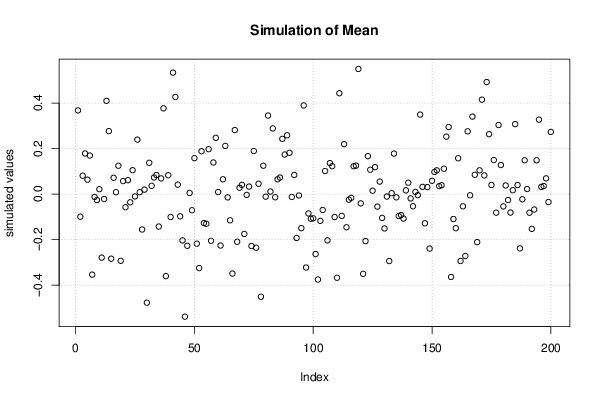
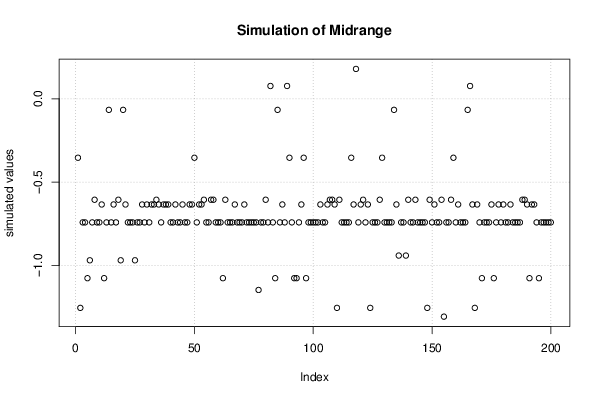
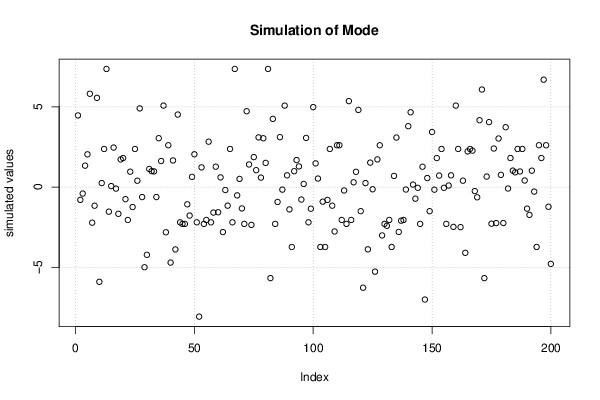
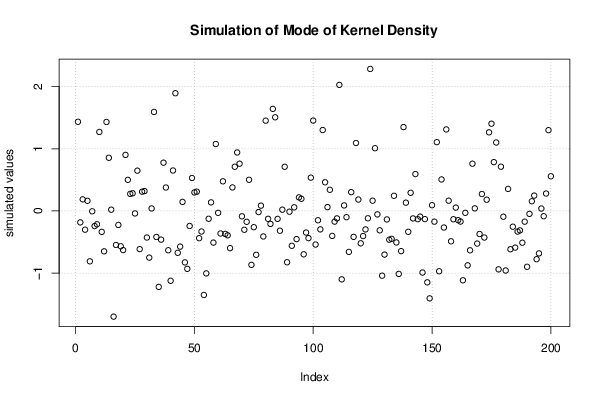
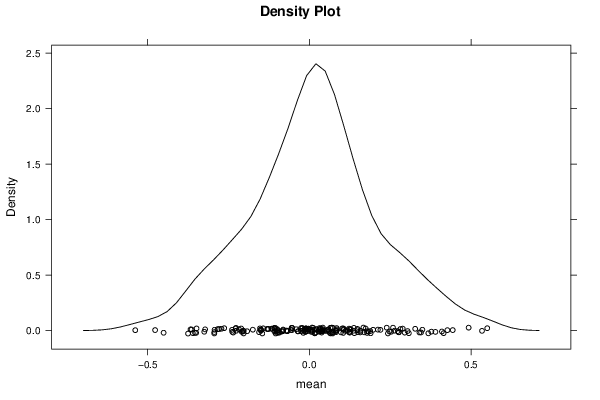
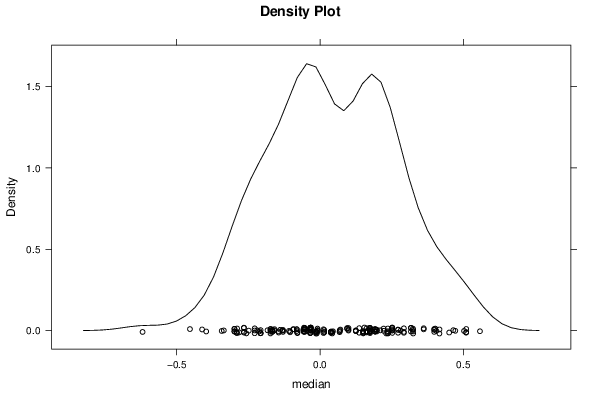
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 200 ; par2 = 5 ; par3 = 0 ; par4 = P1 P5 Q1 Q3 P95 P99 ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par1 <- as.numeric(par1) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||