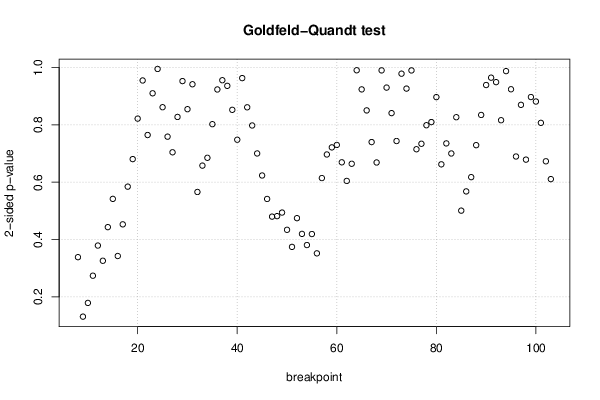
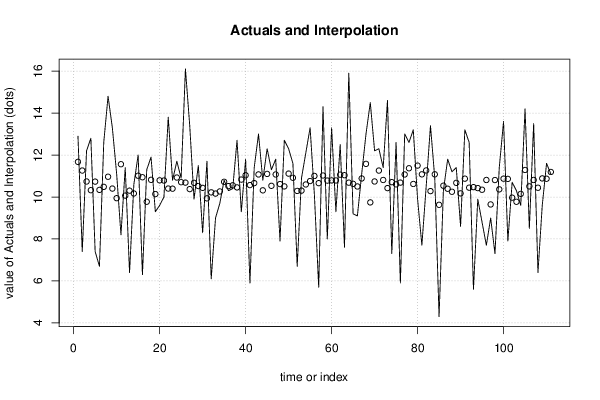
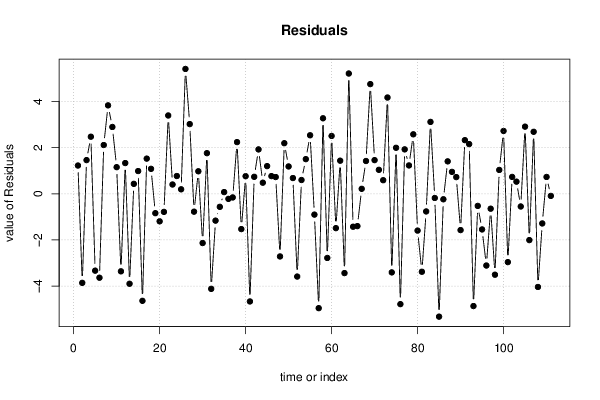
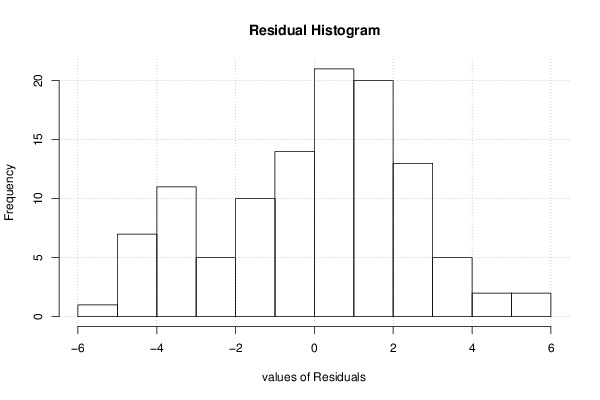
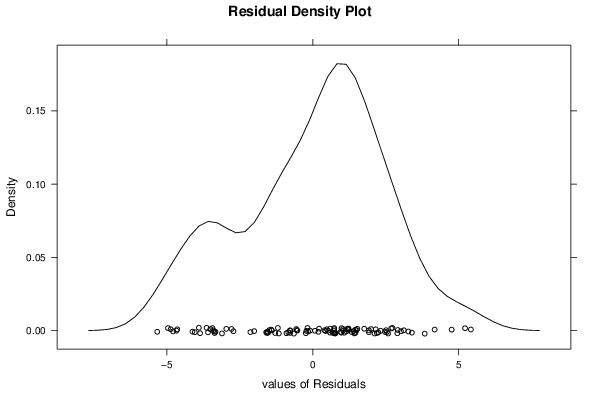
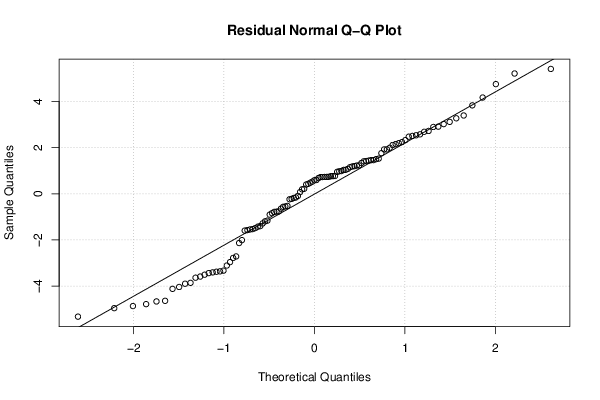
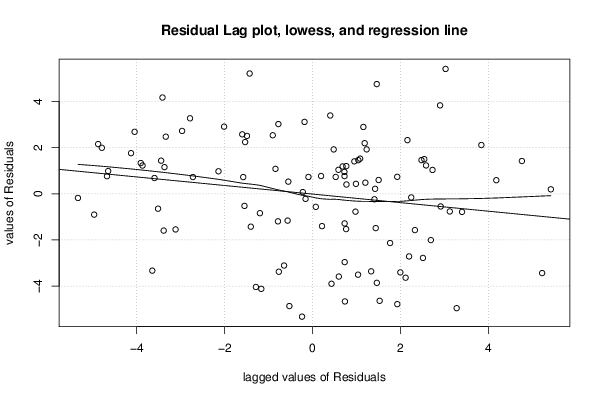
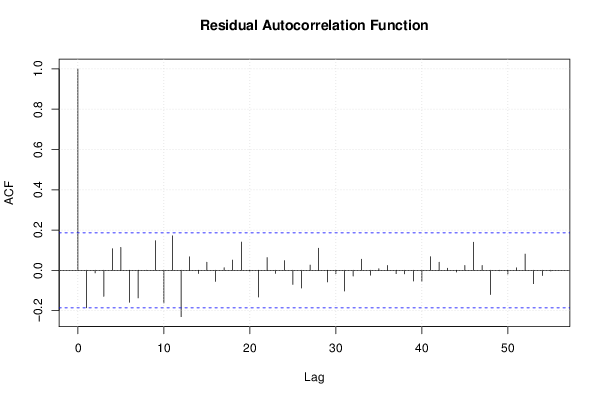
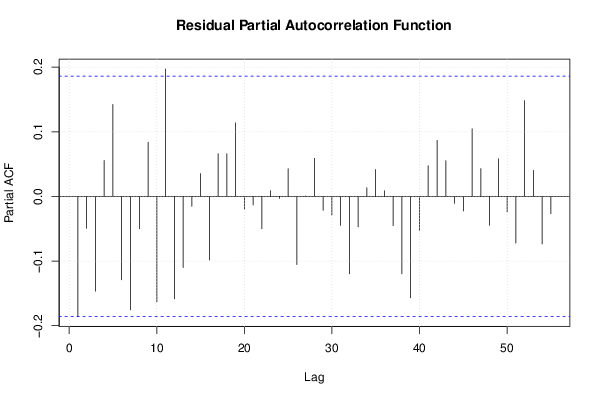
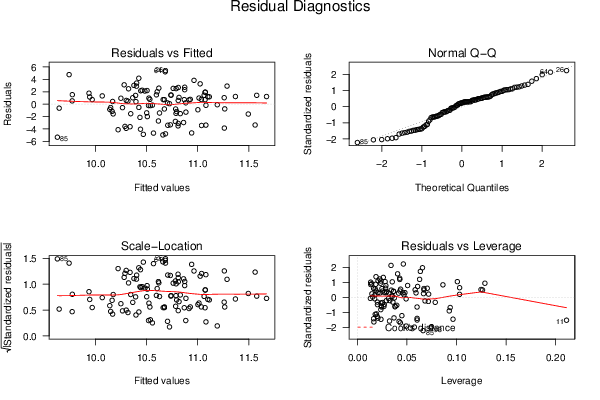
26 50 3,19047619 1,785714286 12,9
51 68 3,476190476 2 7,4
57 62 3,19047619 2,285714286 12,2
37 54 3 3,357142857 12,8
67 71 3,19047619 1,857142857 7,4
43 54 2,761904762 2,714285714 6,7
52 65 3,095238095 2,642857143 12,6
52 73 3,761904762 2,928571429 14,8
43 52 2,571428571 2,285714286 13,3
84 84 3,19047619 2,5 11,1
67 42 3,666666667 1,785714286 8,2
49 66 2,285714286 1,928571429 11,4
70 65 3,095238095 2,428571429 6,4
52 78 2,761904762 2,357142857 10,6
58 73 3,285714286 1,785714286 12,0
68 75 3,857142857 2,642857143 6,3
62 72 3,428571429 4,071428571 11,3
43 66 3 2,142857143 11,9
56 70 3 2,857142857 9,3
56 61 3,333333333 2,5 9,6
74 81 3,666666667 2,285714286 10,0
63 69 3,19047619 2,571428571 13,8
58 71 3,238095238 2,785714286 10,8
63 68 3,666666667 2,571428571 11,7
53 70 3,666666667 3,214285714 10,9
57 68 3,80952381 3,428571429 16,1
51 61 2,714285714 2,214285714 13,4
64 67 3,285714286 2,285714286 9,9
53 76 2,857142857 1,928571429 11,5
29 70 2,19047619 1,642857143 8,3
54 60 2,80952381 3,071428571 11,7
51 77 3,095238095 2,928571429 6,1
58 72 2,761904762 2,285714286 9,0
43 69 3,285714286 3,571428571 9,7
51 71 3,238095238 2,428571429 10,8
53 62 2,80952381 2,071428571 10,3
54 70 3,238095238 2,642857143 10,4
56 64 3,047619048 2,5 12,7
61 58 3,380952381 2,428571429 9,3
47 76 3,428571429 2,285714286 11,8
39 52 2,238095238 1,5 5,9
48 59 2,952380952 2,285714286 11,4
50 68 3,904761905 3,142857143 13,0
35 76 2,19047619 1,571428571 10,8
30 65 3,238095238 2,5 12,3
68 67 3,333333333 2,5 11,3
49 59 3 1,642857143 11,8
61 69 3,047619048 2 7,9
67 76 4,333333333 4,285714286 12,7
47 63 3,619047619 2,714285714 12,3
56 75 3,142857143 1,714285714 11,6
50 63 2,904761905 2,714285714 6,7
43 60 2,047619048 1,357142857 10,9
67 73 3,428571429 2,5 12,1
62 63 2,952380952 1,642857143 13,3
57 70 3,619047619 2,5 10,1
41 75 3,285714286 2,857142857 5,7
54 66 3,095238095 1,642857143 14,3
45 63 2,857142857 1,928571429 8,0
48 63 3,142857143 2,357142857 13,3
61 64 3,047619048 1,785714286 9,3
56 70 3,428571429 2,071428571 12,5
41 75 3,904761905 3,357142857 7,6
43 61 2,523809524 1,571428571 15,9
53 60 3,238095238 2,714285714 9,2
44 62 2,380952381 1,571428571 9,1
66 73 3,523809524 2,214285714 11,1
58 61 3,857142857 2,071428571 13,0
46 66 2,285714286 2,571428571 14,5
37 64 2,238095238 1,071428571 12,2
51 59 3,666666667 2,5 12,3
51 64 3,333333333 2,571428571 11,4
66 56 2,952380952 2,214285714 14,6
45 66 3,238095238 2,714285714 7,3
37 78 2,619047619 1,785714286 12,6
59 53 3,095238095 2,285714286 5,9
42 67 3,19047619 2,071428571 13,0
38 59 3,523809524 2,428571429 12,6
66 66 3,142857143 2,071428571 13,2
34 68 3 1,214285714 9,9
53 71 2,904761905 1,142857143 7,7
49 66 3,238095238 1,642857143 10,5
55 73 2,523809524 1,714285714 13,4
49 72 3,428571429 2,214285714 10,9
59 71 2,428571429 2,571428571 4,3
40 59 3,095238095 3 10,3
58 64 2,904761905 2,285714286 11,8
60 66 2,666666667 2 11,2
63 78 3,904761905 3,285714286 11,4
56 68 2,666666667 2,214285714 8,6
54 73 3,285714286 2,142857143 13,2
52 62 2,761904762 2,142857143 12,6
34 65 3,142857143 3,285714286 5,6
69 68 3,142857143 2,285714286 9,9
32 65 2,380952381 2,142857143 8,8
48 60 2,904761905 1,928571429 7,7
67 71 2,571428571 2,571428571 9,0
58 65 3,142857143 2 7,3
57 68 2,666666667 1,857142857 11,4
42 64 3,095238095 2,285714286 13,6
64 74 3,904761905 3 7,9
58 69 3,095238095 3,285714286 10,7
66 76 2,523809524 2,214285714 10,3
61 72 3,142857143 2,928571429 9,6
52 67 3,238095238 1,5 14,2
51 63 3,238095238 2,928571429 8,5
55 59 3,238095238 2,357142857 13,5
60 66 3,19047619 2,642857143 6,4
56 62 2,80952381 1,357142857 9,6
63 69 3,428571429 2,214285714 11,6
61 66 3,476190476 1,857142857 11,1 |