Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_Tests to Compare Two Means.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | T-Tests | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Thu, 31 May 2012 10:30:47 -0400 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2012/May/31/t1338474668y2spori64ncx9jj.htm/, Retrieved Mon, 06 May 2024 18:46:22 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=168093, Retrieved Mon, 06 May 2024 18:46:22 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 145 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [Histogram, QQplot and Density] [Female weight des...] [2012-05-30 09:46:42] [bee56a896f26c17646228a77f17a2aff] - R D [Histogram, QQplot and Density] [Male weight descr...] [2012-05-30 09:54:00] [bee56a896f26c17646228a77f17a2aff] - RMPD [T-Tests] [Weight and RepWei...] [2012-05-31 12:59:56] [bee56a896f26c17646228a77f17a2aff] - R D [T-Tests] [Weight and RepWei...] [2012-05-31 14:24:15] [bee56a896f26c17646228a77f17a2aff] - D [T-Tests] [Weight and RepWei...] [2012-05-31 14:26:41] [bee56a896f26c17646228a77f17a2aff] - D [T-Tests] [Height and RepHei...] [2012-05-31 14:28:34] [bee56a896f26c17646228a77f17a2aff] - D [T-Tests] [Height and RepHei...] [2012-05-31 14:30:47] [c60080e90d514f19ee1378b89a456a2a] [Current] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
169 165 168 170 163 161 173 170 182 183 170 170 178 175 168 168 178 175 176 175 171 170 175 173 187 188 176 172 178 178 173 170 173 175 175 175 170 165 179 179 177 175 165 165 174 173 186 180 174 171 182 180 183 183 180 180 167 165 172 174 175 174 173 170 173 173 178 175 177 170 180 175 178 178 183 180 169 170 175 175 178 175 172 169 169 165 170 165 167 165 197 200 169 165 183 180 182 180 183 180 178 175 173 173 179 171 177 178 178 178 178 175 176 175 178 175 182 183 184 181 177 175 180 180 184 183 183 183 179 175 191 188 185 185 178 175 189 185 184 183 185 188 173 173 188 185 181 178 187 185 184 183 191 188 189 185 183 180 185 185 185 182 180 178 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
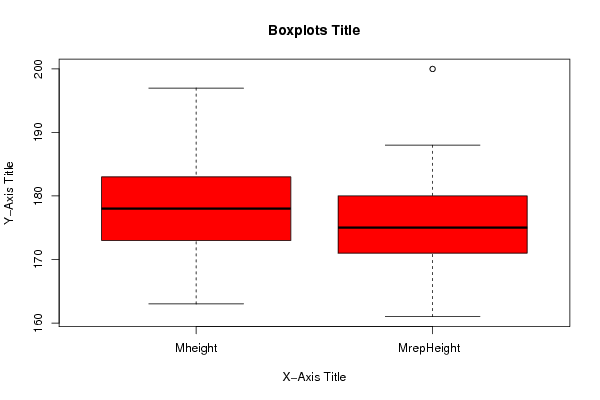
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = two.sided ; par2 = 1 ; par3 = 2 ; par4 = T-Test ; par5 = paired ; par6 = 0.0 ; par7 = 0.95 ; par8 = TRUE ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = two.sided ; par2 = 1 ; par3 = 2 ; par4 = T-Test ; par5 = paired ; par6 = 0.0 ; par7 = 0.95 ; par8 = TRUE ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par2 <- as.numeric(par2) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||