par2 <- as.numeric(par2)
par3 <- as.numeric(par3)
par4 <- as.character(par4)
par5 <- as.character(par5)
par6 <- as.numeric(par6)
par7 <- as.numeric(par7)
par8 <- as.logical(par8)
if ( par5 == 'unpaired') paired <- FALSE else paired <- TRUE
x <- t(y)
if(par8){
bitmap(file='test1.png')
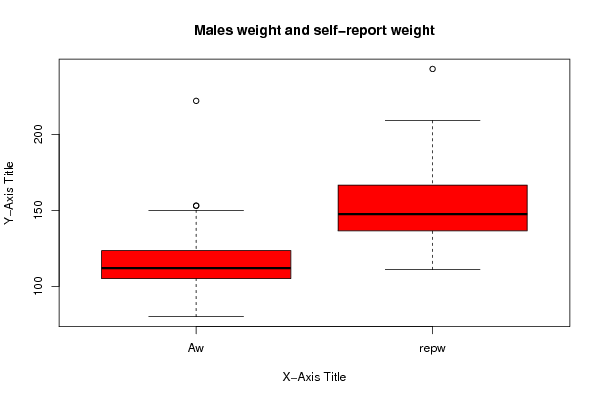
(r<-boxplot(x ,xlab=xlab,ylab=ylab,main=main,notch=FALSE,col=2))
dev.off()
}
load(file='createtable')
if( par4 == 'Wilcoxon-Mann_Whitney'){
a<-table.start()
a <- table.row.start(a)
a <- table.element(a,'Wilcoxon Test',3,TRUE)
a <- table.row.end(a)
a <- table.row.start(a)
a <- table.element(a,'',1,TRUE)
a <- table.element(a,'Statistic',1,TRUE)
a <- table.element(a,'P-value',1,TRUE)
a <- table.row.end(a)
W <- wilcox.test(x[,par2],x[,par3],alternative=par1, paired = paired)
a<-table.row.start(a)
a<-table.element(a,'Wilcoxon Test',1,TRUE)
a<-table.element(a,W$statistic[[1]])
a<-table.element(a,round(W$p.value, digits=5) )
a<-table.row.end(a)
a<-table.end(a)
table.save(a,file='mytable.tab')
}
if( par4 == 'T-Test')
{
T <- t.test(x[,par2],x[,par3],alternative=par1, paired=paired, mu=par6, conf.level=par7)
a<-table.start()
a <- table.row.start(a)
a <- table.element(a,'T-Test',3,TRUE)
a <- table.row.end(a)
if(paired){
a <- table.row.start(a)
a <- table.element(a,'Difference: Mean1 - Mean2',1,TRUE)
a<-table.element(a,round(T$estimate, digits=5) )
a <- table.row.end(a)
}
if(!paired){
a <- table.row.start(a)
a <- table.element(a,'Mean1',1,TRUE)
a<-table.element(a,round(T$estimate[1], digits=5) )
a <- table.row.end(a)
a <- table.row.start(a)
a <- table.element(a,'Mean2',1,TRUE)
a<-table.element(a,round(T$estimate[2], digits=5) )
a <- table.row.end(a)
}
a <- table.row.start(a)
a <- table.element(a,'T Statistic',1,TRUE)
a<-table.element(a,round(T$statistic, digits=5) )
a <- table.row.end(a)
a <- table.row.start(a)
a <- table.element(a,'P-value',1,TRUE)
a<-table.element(a,round(T$p.value, digits=5) )
a <- table.row.end(a)
a <- table.row.start(a)
a <- table.element(a,'Lower Confidence Limit',1,TRUE)
a<-table.element(a,round(T$conf.int[1], digits=5) )
a <- table.row.end(a)
a<-table.row.start(a)
a <- table.element(a,'Upper Confidence Limit',1,TRUE)
a<-table.element(a,round(T$conf.int[2], digits=5) )
a <- table.row.end(a)
a<-table.end(a)
table.save(a,file='mytable.tab')
}
a<-table.start()
a <- table.row.start(a)
a <- table.element(a,'Standard Deviations',3,TRUE)
a <- table.row.end(a)
a <- table.row.start(a)
a <- table.element(a,'Variable 1',1,TRUE)
a<-table.element(a,round(sd(x[,par2], na.rm=TRUE), digits=5) )
a <- table.row.end(a)
a <- table.row.start(a)
a <- table.element(a,'Variable 2',1,TRUE)
a<-table.element(a,round(sd(x[,par3], na.rm=TRUE), digits=5) )
a <- table.row.end(a)
a<-table.end(a)
table.save(a,file='mytable1.tab')
|