\begin{tabular}{lllllllll}
\hline
Summary of computational transaction \tabularnewline
Raw Input & view raw input (R code) \tabularnewline
Raw Output & view raw output of R engine \tabularnewline
Computing time & 1 seconds \tabularnewline
R Server & 'Gertrude Mary Cox' @ cox.wessa.net \tabularnewline
\hline
\end{tabular}
%Source: https://freestatistics.org/blog/index.php?pk=162335&T=0
[TABLE]
[ROW][C]Summary of computational transaction[/C][/ROW]
[ROW][C]Raw Input[/C][C]view raw input (R code) [/C][/ROW]
[ROW][C]Raw Output[/C][C]view raw output of R engine [/C][/ROW]
[ROW][C]Computing time[/C][C]1 seconds[/C][/ROW]
[ROW][C]R Server[/C][C]'Gertrude Mary Cox' @ cox.wessa.net[/C][/ROW]
[/TABLE]
Source: https://freestatistics.org/blog/index.php?pk=162335&T=0
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
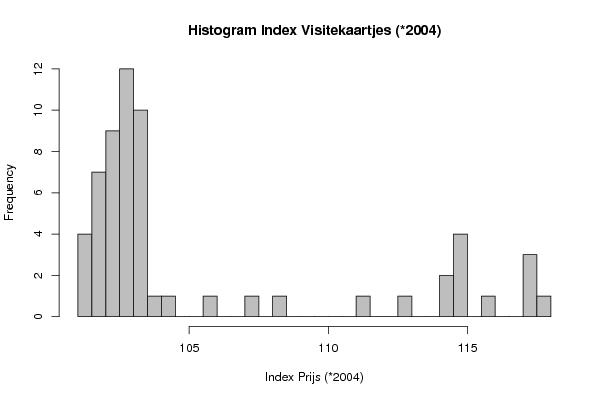
| Frequency Table (Histogram) | | Bins | Midpoint | Abs. Frequency | Rel. Frequency | Cumul. Rel. Freq. | Density | | [101,101.5[ | 101.25 | 4 | 0.066667 | 0.066667 | 0.133333 | | [101.5,102[ | 101.75 | 7 | 0.116667 | 0.183333 | 0.233333 | | [102,102.5[ | 102.25 | 9 | 0.15 | 0.333333 | 0.3 | | [102.5,103[ | 102.75 | 12 | 0.2 | 0.533333 | 0.4 | | [103,103.5[ | 103.25 | 10 | 0.166667 | 0.7 | 0.333333 | | [103.5,104[ | 103.75 | 1 | 0.016667 | 0.716667 | 0.033333 | | [104,104.5[ | 104.25 | 1 | 0.016667 | 0.733333 | 0.033333 | | [104.5,105[ | 104.75 | 0 | 0 | 0.733333 | 0 | | [105,105.5[ | 105.25 | 0 | 0 | 0.733333 | 0 | | [105.5,106[ | 105.75 | 1 | 0.016667 | 0.75 | 0.033333 | | [106,106.5[ | 106.25 | 0 | 0 | 0.75 | 0 | | [106.5,107[ | 106.75 | 0 | 0 | 0.75 | 0 | | [107,107.5[ | 107.25 | 1 | 0.016667 | 0.766667 | 0.033333 | | [107.5,108[ | 107.75 | 0 | 0 | 0.766667 | 0 | | [108,108.5[ | 108.25 | 1 | 0.016667 | 0.783333 | 0.033333 | | [108.5,109[ | 108.75 | 0 | 0 | 0.783333 | 0 | | [109,109.5[ | 109.25 | 0 | 0 | 0.783333 | 0 | | [109.5,110[ | 109.75 | 0 | 0 | 0.783333 | 0 | | [110,110.5[ | 110.25 | 0 | 0 | 0.783333 | 0 | | [110.5,111[ | 110.75 | 0 | 0 | 0.783333 | 0 | | [111,111.5[ | 111.25 | 1 | 0.016667 | 0.8 | 0.033333 | | [111.5,112[ | 111.75 | 0 | 0 | 0.8 | 0 | | [112,112.5[ | 112.25 | 0 | 0 | 0.8 | 0 | | [112.5,113[ | 112.75 | 1 | 0.016667 | 0.816667 | 0.033333 | | [113,113.5[ | 113.25 | 0 | 0 | 0.816667 | 0 | | [113.5,114[ | 113.75 | 0 | 0 | 0.816667 | 0 | | [114,114.5[ | 114.25 | 2 | 0.033333 | 0.85 | 0.066667 | | [114.5,115[ | 114.75 | 4 | 0.066667 | 0.916667 | 0.133333 | | [115,115.5[ | 115.25 | 0 | 0 | 0.916667 | 0 | | [115.5,116[ | 115.75 | 1 | 0.016667 | 0.933333 | 0.033333 | | [116,116.5[ | 116.25 | 0 | 0 | 0.933333 | 0 | | [116.5,117[ | 116.75 | 0 | 0 | 0.933333 | 0 | | [117,117.5[ | 117.25 | 3 | 0.05 | 0.983333 | 0.1 | | [117.5,118] | 117.75 | 1 | 0.016667 | 1 | 0.033333 |
\begin{tabular}{lllllllll}
\hline
Frequency Table (Histogram) \tabularnewline
Bins & Midpoint & Abs. Frequency & Rel. Frequency & Cumul. Rel. Freq. & Density \tabularnewline
[101,101.5[ & 101.25 & 4 & 0.066667 & 0.066667 & 0.133333 \tabularnewline
[101.5,102[ & 101.75 & 7 & 0.116667 & 0.183333 & 0.233333 \tabularnewline
[102,102.5[ & 102.25 & 9 & 0.15 & 0.333333 & 0.3 \tabularnewline
[102.5,103[ & 102.75 & 12 & 0.2 & 0.533333 & 0.4 \tabularnewline
[103,103.5[ & 103.25 & 10 & 0.166667 & 0.7 & 0.333333 \tabularnewline
[103.5,104[ & 103.75 & 1 & 0.016667 & 0.716667 & 0.033333 \tabularnewline
[104,104.5[ & 104.25 & 1 & 0.016667 & 0.733333 & 0.033333 \tabularnewline
[104.5,105[ & 104.75 & 0 & 0 & 0.733333 & 0 \tabularnewline
[105,105.5[ & 105.25 & 0 & 0 & 0.733333 & 0 \tabularnewline
[105.5,106[ & 105.75 & 1 & 0.016667 & 0.75 & 0.033333 \tabularnewline
[106,106.5[ & 106.25 & 0 & 0 & 0.75 & 0 \tabularnewline
[106.5,107[ & 106.75 & 0 & 0 & 0.75 & 0 \tabularnewline
[107,107.5[ & 107.25 & 1 & 0.016667 & 0.766667 & 0.033333 \tabularnewline
[107.5,108[ & 107.75 & 0 & 0 & 0.766667 & 0 \tabularnewline
[108,108.5[ & 108.25 & 1 & 0.016667 & 0.783333 & 0.033333 \tabularnewline
[108.5,109[ & 108.75 & 0 & 0 & 0.783333 & 0 \tabularnewline
[109,109.5[ & 109.25 & 0 & 0 & 0.783333 & 0 \tabularnewline
[109.5,110[ & 109.75 & 0 & 0 & 0.783333 & 0 \tabularnewline
[110,110.5[ & 110.25 & 0 & 0 & 0.783333 & 0 \tabularnewline
[110.5,111[ & 110.75 & 0 & 0 & 0.783333 & 0 \tabularnewline
[111,111.5[ & 111.25 & 1 & 0.016667 & 0.8 & 0.033333 \tabularnewline
[111.5,112[ & 111.75 & 0 & 0 & 0.8 & 0 \tabularnewline
[112,112.5[ & 112.25 & 0 & 0 & 0.8 & 0 \tabularnewline
[112.5,113[ & 112.75 & 1 & 0.016667 & 0.816667 & 0.033333 \tabularnewline
[113,113.5[ & 113.25 & 0 & 0 & 0.816667 & 0 \tabularnewline
[113.5,114[ & 113.75 & 0 & 0 & 0.816667 & 0 \tabularnewline
[114,114.5[ & 114.25 & 2 & 0.033333 & 0.85 & 0.066667 \tabularnewline
[114.5,115[ & 114.75 & 4 & 0.066667 & 0.916667 & 0.133333 \tabularnewline
[115,115.5[ & 115.25 & 0 & 0 & 0.916667 & 0 \tabularnewline
[115.5,116[ & 115.75 & 1 & 0.016667 & 0.933333 & 0.033333 \tabularnewline
[116,116.5[ & 116.25 & 0 & 0 & 0.933333 & 0 \tabularnewline
[116.5,117[ & 116.75 & 0 & 0 & 0.933333 & 0 \tabularnewline
[117,117.5[ & 117.25 & 3 & 0.05 & 0.983333 & 0.1 \tabularnewline
[117.5,118] & 117.75 & 1 & 0.016667 & 1 & 0.033333 \tabularnewline
\hline
\end{tabular}
%Source: https://freestatistics.org/blog/index.php?pk=162335&T=1
[TABLE]
[ROW][C]Frequency Table (Histogram)[/C][/ROW]
[ROW][C]Bins[/C][C]Midpoint[/C][C]Abs. Frequency[/C][C]Rel. Frequency[/C][C]Cumul. Rel. Freq.[/C][C]Density[/C][/ROW]
[ROW][C][101,101.5[[/C][C]101.25[/C][C]4[/C][C]0.066667[/C][C]0.066667[/C][C]0.133333[/C][/ROW]
[ROW][C][101.5,102[[/C][C]101.75[/C][C]7[/C][C]0.116667[/C][C]0.183333[/C][C]0.233333[/C][/ROW]
[ROW][C][102,102.5[[/C][C]102.25[/C][C]9[/C][C]0.15[/C][C]0.333333[/C][C]0.3[/C][/ROW]
[ROW][C][102.5,103[[/C][C]102.75[/C][C]12[/C][C]0.2[/C][C]0.533333[/C][C]0.4[/C][/ROW]
[ROW][C][103,103.5[[/C][C]103.25[/C][C]10[/C][C]0.166667[/C][C]0.7[/C][C]0.333333[/C][/ROW]
[ROW][C][103.5,104[[/C][C]103.75[/C][C]1[/C][C]0.016667[/C][C]0.716667[/C][C]0.033333[/C][/ROW]
[ROW][C][104,104.5[[/C][C]104.25[/C][C]1[/C][C]0.016667[/C][C]0.733333[/C][C]0.033333[/C][/ROW]
[ROW][C][104.5,105[[/C][C]104.75[/C][C]0[/C][C]0[/C][C]0.733333[/C][C]0[/C][/ROW]
[ROW][C][105,105.5[[/C][C]105.25[/C][C]0[/C][C]0[/C][C]0.733333[/C][C]0[/C][/ROW]
[ROW][C][105.5,106[[/C][C]105.75[/C][C]1[/C][C]0.016667[/C][C]0.75[/C][C]0.033333[/C][/ROW]
[ROW][C][106,106.5[[/C][C]106.25[/C][C]0[/C][C]0[/C][C]0.75[/C][C]0[/C][/ROW]
[ROW][C][106.5,107[[/C][C]106.75[/C][C]0[/C][C]0[/C][C]0.75[/C][C]0[/C][/ROW]
[ROW][C][107,107.5[[/C][C]107.25[/C][C]1[/C][C]0.016667[/C][C]0.766667[/C][C]0.033333[/C][/ROW]
[ROW][C][107.5,108[[/C][C]107.75[/C][C]0[/C][C]0[/C][C]0.766667[/C][C]0[/C][/ROW]
[ROW][C][108,108.5[[/C][C]108.25[/C][C]1[/C][C]0.016667[/C][C]0.783333[/C][C]0.033333[/C][/ROW]
[ROW][C][108.5,109[[/C][C]108.75[/C][C]0[/C][C]0[/C][C]0.783333[/C][C]0[/C][/ROW]
[ROW][C][109,109.5[[/C][C]109.25[/C][C]0[/C][C]0[/C][C]0.783333[/C][C]0[/C][/ROW]
[ROW][C][109.5,110[[/C][C]109.75[/C][C]0[/C][C]0[/C][C]0.783333[/C][C]0[/C][/ROW]
[ROW][C][110,110.5[[/C][C]110.25[/C][C]0[/C][C]0[/C][C]0.783333[/C][C]0[/C][/ROW]
[ROW][C][110.5,111[[/C][C]110.75[/C][C]0[/C][C]0[/C][C]0.783333[/C][C]0[/C][/ROW]
[ROW][C][111,111.5[[/C][C]111.25[/C][C]1[/C][C]0.016667[/C][C]0.8[/C][C]0.033333[/C][/ROW]
[ROW][C][111.5,112[[/C][C]111.75[/C][C]0[/C][C]0[/C][C]0.8[/C][C]0[/C][/ROW]
[ROW][C][112,112.5[[/C][C]112.25[/C][C]0[/C][C]0[/C][C]0.8[/C][C]0[/C][/ROW]
[ROW][C][112.5,113[[/C][C]112.75[/C][C]1[/C][C]0.016667[/C][C]0.816667[/C][C]0.033333[/C][/ROW]
[ROW][C][113,113.5[[/C][C]113.25[/C][C]0[/C][C]0[/C][C]0.816667[/C][C]0[/C][/ROW]
[ROW][C][113.5,114[[/C][C]113.75[/C][C]0[/C][C]0[/C][C]0.816667[/C][C]0[/C][/ROW]
[ROW][C][114,114.5[[/C][C]114.25[/C][C]2[/C][C]0.033333[/C][C]0.85[/C][C]0.066667[/C][/ROW]
[ROW][C][114.5,115[[/C][C]114.75[/C][C]4[/C][C]0.066667[/C][C]0.916667[/C][C]0.133333[/C][/ROW]
[ROW][C][115,115.5[[/C][C]115.25[/C][C]0[/C][C]0[/C][C]0.916667[/C][C]0[/C][/ROW]
[ROW][C][115.5,116[[/C][C]115.75[/C][C]1[/C][C]0.016667[/C][C]0.933333[/C][C]0.033333[/C][/ROW]
[ROW][C][116,116.5[[/C][C]116.25[/C][C]0[/C][C]0[/C][C]0.933333[/C][C]0[/C][/ROW]
[ROW][C][116.5,117[[/C][C]116.75[/C][C]0[/C][C]0[/C][C]0.933333[/C][C]0[/C][/ROW]
[ROW][C][117,117.5[[/C][C]117.25[/C][C]3[/C][C]0.05[/C][C]0.983333[/C][C]0.1[/C][/ROW]
[ROW][C][117.5,118][/C][C]117.75[/C][C]1[/C][C]0.016667[/C][C]1[/C][C]0.033333[/C][/ROW]
[/TABLE]
Source: https://freestatistics.org/blog/index.php?pk=162335&T=1
Globally Unique Identifier (entire table): ba.freestatistics.org/blog/index.php?pk=162335&T=1
As an alternative you can also use a QR Code:
The GUIDs for individual cells are displayed in the table below:
| Frequency Table (Histogram) | | Bins | Midpoint | Abs. Frequency | Rel. Frequency | Cumul. Rel. Freq. | Density | | [101,101.5[ | 101.25 | 4 | 0.066667 | 0.066667 | 0.133333 | | [101.5,102[ | 101.75 | 7 | 0.116667 | 0.183333 | 0.233333 | | [102,102.5[ | 102.25 | 9 | 0.15 | 0.333333 | 0.3 | | [102.5,103[ | 102.75 | 12 | 0.2 | 0.533333 | 0.4 | | [103,103.5[ | 103.25 | 10 | 0.166667 | 0.7 | 0.333333 | | [103.5,104[ | 103.75 | 1 | 0.016667 | 0.716667 | 0.033333 | | [104,104.5[ | 104.25 | 1 | 0.016667 | 0.733333 | 0.033333 | | [104.5,105[ | 104.75 | 0 | 0 | 0.733333 | 0 | | [105,105.5[ | 105.25 | 0 | 0 | 0.733333 | 0 | | [105.5,106[ | 105.75 | 1 | 0.016667 | 0.75 | 0.033333 | | [106,106.5[ | 106.25 | 0 | 0 | 0.75 | 0 | | [106.5,107[ | 106.75 | 0 | 0 | 0.75 | 0 | | [107,107.5[ | 107.25 | 1 | 0.016667 | 0.766667 | 0.033333 | | [107.5,108[ | 107.75 | 0 | 0 | 0.766667 | 0 | | [108,108.5[ | 108.25 | 1 | 0.016667 | 0.783333 | 0.033333 | | [108.5,109[ | 108.75 | 0 | 0 | 0.783333 | 0 | | [109,109.5[ | 109.25 | 0 | 0 | 0.783333 | 0 | | [109.5,110[ | 109.75 | 0 | 0 | 0.783333 | 0 | | [110,110.5[ | 110.25 | 0 | 0 | 0.783333 | 0 | | [110.5,111[ | 110.75 | 0 | 0 | 0.783333 | 0 | | [111,111.5[ | 111.25 | 1 | 0.016667 | 0.8 | 0.033333 | | [111.5,112[ | 111.75 | 0 | 0 | 0.8 | 0 | | [112,112.5[ | 112.25 | 0 | 0 | 0.8 | 0 | | [112.5,113[ | 112.75 | 1 | 0.016667 | 0.816667 | 0.033333 | | [113,113.5[ | 113.25 | 0 | 0 | 0.816667 | 0 | | [113.5,114[ | 113.75 | 0 | 0 | 0.816667 | 0 | | [114,114.5[ | 114.25 | 2 | 0.033333 | 0.85 | 0.066667 | | [114.5,115[ | 114.75 | 4 | 0.066667 | 0.916667 | 0.133333 | | [115,115.5[ | 115.25 | 0 | 0 | 0.916667 | 0 | | [115.5,116[ | 115.75 | 1 | 0.016667 | 0.933333 | 0.033333 | | [116,116.5[ | 116.25 | 0 | 0 | 0.933333 | 0 | | [116.5,117[ | 116.75 | 0 | 0 | 0.933333 | 0 | | [117,117.5[ | 117.25 | 3 | 0.05 | 0.983333 | 0.1 | | [117.5,118] | 117.75 | 1 | 0.016667 | 1 | 0.033333 |
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
If you paste this QR Code into your document, anyone with a smartphone or tablet will be able to scan it and view this table in a browser.
|