Free Statistics
of Irreproducible Research!
Description of Statistical Computation | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | ||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | |||||||||||||||||||||||||||||
| R Software Module | rwasp_Distributional Plots.wasp | |||||||||||||||||||||||||||||
| Title produced by software | Histogram, QQplot and Density | |||||||||||||||||||||||||||||
| Date of computation | Tue, 14 Feb 2012 13:01:42 -0500 | |||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2012/Feb/14/t1329242571oaf658afifadw04.htm/, Retrieved Mon, 29 Apr 2024 10:11:33 +0000 | |||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=162326, Retrieved Mon, 29 Apr 2024 10:11:33 +0000 | ||||||||||||||||||||||||||||||
| QR Codes: | ||||||||||||||||||||||||||||||
|
| ||||||||||||||||||||||||||||||
| Original text written by user: | ||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | |||||||||||||||||||||||||||||
| User-defined keywords | ||||||||||||||||||||||||||||||
| Estimated Impact | 258 | |||||||||||||||||||||||||||||
Tree of Dependent Computations | ||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | ||||||||||||||||||||||||||||||
| - [T-Tests] [Salk Heatbeat Study] [2012-02-14 16:56:38] [98fd0e87c3eb04e0cc2efde01dbafab6] - RMPD [Histogram, QQplot and Density] [Salk Heatbeat Stu...] [2012-02-14 18:01:42] [a9208f4f8d3b118336aae915785f2bd9] [Current] - RMP [Histogram, QQplot and Density] [heartbeat] [2012-05-03 09:23:27] [ea6e45d26e44fca6fcec5c603ff1140c] - RMP [Histogram, QQplot and Density] [grpahs for heartb...] [2012-05-03 09:23:50] [c2d7eae68f5ec0337d2d2ba826377ba0] - RMP [Histogram, QQplot and Density] [] [2012-05-03 09:31:15] [2d5cef47d6186b035b8ade2ed171f642] - R P [Histogram, QQplot and Density] [heartbeat] [2012-05-03 09:31:12] [35521cdb85a31d9f6eb48e92142bc928] - R P [Histogram, QQplot and Density] [HB] [2012-05-03 09:32:36] [5af04e13c06bbf1f7fb207eb9550d664] - RMP [Histogram, QQplot and Density] [nheartbeat] [2012-05-03 09:34:44] [c2d7eae68f5ec0337d2d2ba826377ba0] - RMP [Histogram, QQplot and Density] [with heartbeat] [2012-05-03 09:39:00] [dcbd67829fb198dc024d653d5871c807] - R P [Histogram, QQplot and Density] [Graph two] [2012-05-03 09:39:53] [57084a6890c0bc671c16163e98194e4e] - R PD [Histogram, QQplot and Density] [20 bins and 10% t...] [2012-05-03 09:39:54] [bf1ddb3cd45fb5d723b760bc40755ff6] - R P [Histogram, QQplot and Density] [Babies exposed to...] [2012-05-03 09:41:34] [b7df83e0fc4d05ddde076a5a0ec0675f] - RMP [Histogram, QQplot and Density] [Distributional pl...] [2012-05-03 09:41:45] [a68359a26e24fd92b609f4c95c254176] - RMP [Histogram, QQplot and Density] [] [2012-05-03 09:42:39] [a63c53a5ffd62668590cfb76aad837c1] - R PD [Histogram, QQplot and Density] [all data] [2012-05-03 09:43:33] [bf1ddb3cd45fb5d723b760bc40755ff6] - RMPD [Wilcoxon-Mann-Whitney Test] [results] [2012-05-03 09:43:40] [dcbd67829fb198dc024d653d5871c807] - RMPD [Wilcoxon-Mann-Whitney Test] [Babies exposed an...] [2012-05-03 09:46:19] [b7df83e0fc4d05ddde076a5a0ec0675f] - RMP [Histogram, QQplot and Density] [] [2012-05-03 09:47:19] [221b3b4216f0c3837d81a9a9f89ff273] - R P [Histogram, QQplot and Density] [Descriptive Stati...] [2012-05-03 09:48:26] [09253b89c68efd7a460a267273a9d6e3] - RMP [Histogram and QQplot] [Heartbeat group] [2012-05-03 09:49:14] [74be16979710d4c4e7c6647856088456] - R P [Histogram, QQplot and Density] [Heartbeat condition] [2012-05-03 09:50:01] [5ab72825a06ca2a73c828d10c85d14d8] - RMPD [T-Tests] [T-test] [2012-05-03 09:50:49] [bf1ddb3cd45fb5d723b760bc40755ff6] - RM [One-Way-Between-Groups ANOVA- Free Statistics Software (Calculator)] [Tukeys] [2012-05-03 10:02:05] [bf1ddb3cd45fb5d723b760bc40755ff6] - R P [Histogram, QQplot and Density] [Heartbeat] [2012-05-03 09:50:54] [1bd8d3e7b11d3e986b30388f410fa8ef] - R P [Histogram, QQplot and Density] [] [2012-05-03 09:54:00] [a7ea7f4263337445a0a4c6547fb2278a] - RMP [Histogram, QQplot and Density] [Heartbeat babies ...] [2012-05-03 09:57:33] [069c1c4384e883dd4e1df6d4e8f70f1e] - RMPD [MC Mock_Exam - Q1] [] [2012-05-03 10:01:41] [9a61272a736950814aceafb9bfc5e47e] - RMP [Histogram, QQplot and Density] [qqplot 2] [2012-08-30 09:48:59] [74be16979710d4c4e7c6647856088456] - R P [Histogram, QQplot and Density] [] [2012-08-30 10:00:56] [e5cd290daa2365fa501ea04dbf3bfa28] - RMPD [Wilcoxon-Mann-Whitney Test] [Wilcoxon Test Hea...] [2013-05-09 09:27:49] [fc21a227dc348c95c639a44f4159bb1c] - RMP [Histogram, QQplot and Density] [] [2013-05-09 09:35:44] [74be16979710d4c4e7c6647856088456] - RMPD [Wilcoxon-Mann-Whitney Test] [Soothing effect o...] [2013-05-09 09:39:37] [526987df82e09676a270245976380ec1] - R [Wilcoxon-Mann-Whitney Test] [Soothing effect o...] [2013-05-09 09:43:42] [526987df82e09676a270245976380ec1] - RMPD [T-Tests] [Difference betwee...] [2013-05-09 09:40:17] [1da54e444dc2679e5d9aa3487bd18b49] - RMPD [Correlation] [hjj] [2013-05-09 09:42:19] [55fd393080df5be6479d375e48464127] | ||||||||||||||||||||||||||||||
| Feedback Forum | ||||||||||||||||||||||||||||||
Post a new message | ||||||||||||||||||||||||||||||
Dataset | ||||||||||||||||||||||||||||||
| Dataseries X: | ||||||||||||||||||||||||||||||
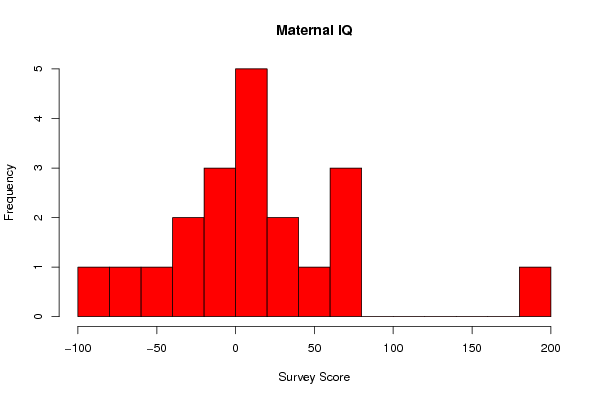
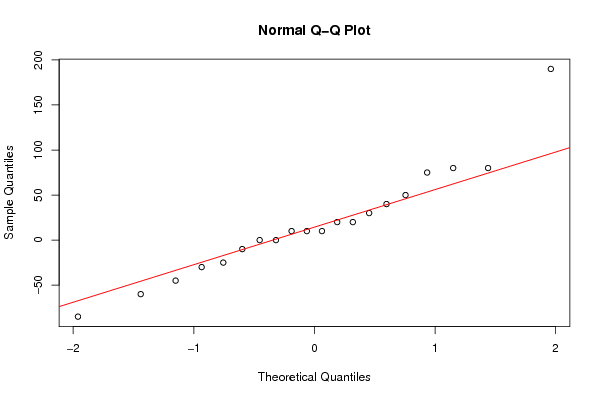
190 80 80 75 50 40 30 20 20 10 10 10 0 0 -10 -25 -30 -45 -60 -85 | ||||||||||||||||||||||||||||||
Tables (Output of Computation) | ||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
Figures (Output of Computation) | ||||||||||||||||||||||||||||||
Input Parameters & R Code | ||||||||||||||||||||||||||||||
| Parameters (Session): | ||||||||||||||||||||||||||||||
| par1 = 3 ; par2 = TRUE ; par3 = 0 ; par4 = 1 ; par5 = 2 ; | ||||||||||||||||||||||||||||||
| Parameters (R input): | ||||||||||||||||||||||||||||||
| par1 = 10 ; | ||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | ||||||||||||||||||||||||||||||
bitmap(file='test1.png') | ||||||||||||||||||||||||||||||