Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_Tests to Compare Two Means.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | T-Tests | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Tue, 14 Feb 2012 11:56:38 -0500 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2012/Feb/14/t13292387940gxd5coql7uku3s.htm/, Retrieved Mon, 29 Apr 2024 14:30:46 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=162316, Retrieved Mon, 29 Apr 2024 14:30:46 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 166 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [T-Tests] [Salk Heatbeat Study] [2012-02-14 16:56:38] [a9208f4f8d3b118336aae915785f2bd9] [Current] - R [T-Tests] [Salk Heatbeat Study] [2012-02-14 17:06:34] [98fd0e87c3eb04e0cc2efde01dbafab6] - RMPD [Histogram, QQplot and Density] [Salk Heatbeat Stu...] [2012-02-14 17:58:15] [98fd0e87c3eb04e0cc2efde01dbafab6] - RMP [Histogram, QQplot and Density] [qq plot] [2012-05-03 09:19:25] [ea6e45d26e44fca6fcec5c603ff1140c] - R P [Histogram, QQplot and Density] [No heartbeat ] [2012-05-03 09:19:54] [35521cdb85a31d9f6eb48e92142bc928] - R P [Histogram, QQplot and Density] [NoHBSample] [2012-05-03 09:21:32] [5af04e13c06bbf1f7fb207eb9550d664] - [Histogram, QQplot and Density] [NoHB] [2012-05-03 09:25:27] [5af04e13c06bbf1f7fb207eb9550d664] - RMP [Histogram, QQplot and Density] [qqplot] [2012-05-03 09:21:53] [c2d7eae68f5ec0337d2d2ba826377ba0] - RMP [Histogram, QQplot and Density] [grpahs for no hea...] [2012-05-03 09:23:04] [c2d7eae68f5ec0337d2d2ba826377ba0] - R P [Histogram, QQplot and Density] [no heartbeat] [2012-05-03 09:25:25] [35521cdb85a31d9f6eb48e92142bc928] - R P [Histogram, QQplot and Density] [] [2012-05-03 09:26:08] [2d5cef47d6186b035b8ade2ed171f642] - RMP [Histogram, QQplot and Density] [] [2012-05-03 09:29:38] [2d5cef47d6186b035b8ade2ed171f642] - RMP [Histogram and QQplot] [] [2012-05-03 09:29:58] [74be16979710d4c4e7c6647856088456] - RMP [Histogram, QQplot and Density] [no heartbeat] [2012-05-03 09:33:10] [c2d7eae68f5ec0337d2d2ba826377ba0] - R P [Histogram, QQplot and Density] [20 bins ] [2012-05-03 09:35:40] [bf1ddb3cd45fb5d723b760bc40755ff6] - R P [Histogram, QQplot and Density] [Graph one] [2012-05-03 09:38:03] [57084a6890c0bc671c16163e98194e4e] - RMP [Histogram, QQplot and Density] [no heartbeat] [2012-05-03 09:37:41] [dcbd67829fb198dc024d653d5871c807] - RMP [Histogram, QQplot and Density] [Box Plot 1 ] [2012-05-03 09:39:21] [f5d7ef0e9b68469df85069f757ece0ab] - R P [Histogram, QQplot and Density] [No heartbeat] [2012-05-03 09:39:22] [1bd8d3e7b11d3e986b30388f410fa8ef] - R P [Histogram, QQplot and Density] [Babies not expose...] [2012-05-03 09:40:00] [b7df83e0fc4d05ddde076a5a0ec0675f] - RMP [Histogram, QQplot and Density] [Plots For No Hear...] [2012-05-03 09:40:01] [9a61272a736950814aceafb9bfc5e47e] - RMP [Histogram, QQplot and Density] [Distributional pl...] [2012-05-03 09:39:58] [a68359a26e24fd92b609f4c95c254176] - RMP [Histogram, QQplot and Density] [Descriptive Stati...] [2012-05-03 09:40:19] [09253b89c68efd7a460a267273a9d6e3] - RMP [Histogram, QQplot and Density] [] [2012-05-03 09:41:11] [a63c53a5ffd62668590cfb76aad837c1] - R P [Histogram, QQplot and Density] [Graph one] [2012-05-03 09:41:22] [57084a6890c0bc671c16163e98194e4e] - RMP [Histogram, QQplot and Density] [Box Plot 1 ] [2012-05-03 09:41:37] [f5d7ef0e9b68469df85069f757ece0ab] - RMP [Histogram, QQplot and Density] [graphs to represe...] [2012-05-03 09:42:34] [04881406e92e7b483917daebdd502463] - RMP [Histogram, QQplot and Density] [Box Plot 1] [2012-05-03 09:42:39] [f5d7ef0e9b68469df85069f757ece0ab] - RMP [Histogram, QQplot and Density] [] [2012-05-03 09:43:55] [221b3b4216f0c3837d81a9a9f89ff273] - RMP [Histogram, QQplot and Density] [] [2012-05-03 09:45:58] [221b3b4216f0c3837d81a9a9f89ff273] - RMP [Histogram, QQplot and Density] [Box Plot 1 ] [2012-05-03 09:47:02] [f5d7ef0e9b68469df85069f757ece0ab] - R P [Histogram, QQplot and Density] [] [2012-05-03 09:48:18] [a7ea7f4263337445a0a4c6547fb2278a] - R P [Histogram, QQplot and Density] [No Heartbeat cond...] [2012-05-03 09:48:27] [5ab72825a06ca2a73c828d10c85d14d8] - RMP [Histogram, QQplot and Density] [Box Plot 1] [2012-05-03 09:48:45] [f5d7ef0e9b68469df85069f757ece0ab] - RMPD [Wilcoxon-Mann-Whitney Test] [Mann whitney test] [2012-05-03 09:49:02] [57084a6890c0bc671c16163e98194e4e] - RMPD [T-Tests] [A comparison of s...] [2012-05-03 09:52:21] [c14255f5a914c4d7aab5f6cd60318f9b] - RMPD [T-Tests] [dgs] [2012-05-03 09:54:25] [9fd651d676601517bf05777b8cc41305] - RMP [Histogram, QQplot and Density] [No heartbeat sample] [2012-05-03 09:56:35] [069c1c4384e883dd4e1df6d4e8f70f1e] - RMPD [MC May Mock - Q6] [] [2012-05-30 23:07:31] [28e3b0ec23e510cd6fa92636dd93aa1d] - RMPD [MC May Mock - Q6] [] [2012-05-30 23:07:31] [28e3b0ec23e510cd6fa92636dd93aa1d] - RMP [Histogram, QQplot and Density] [no heartbeat sample] [2012-06-01 07:09:15] [5934aceae8cf147bda5be230722b868c] - R PD [Histogram, QQplot and Density] [Summer exam] [2012-06-01 09:13:10] [8c387ce24444dc2c0e7d576cfc7658e0] - R PD [Histogram, QQplot and Density] [Summer exam] [2012-06-01 09:16:53] [8c387ce24444dc2c0e7d576cfc7658e0] - RMPD [Wilcoxon-Mann-Whitney Test] [Summer exam] [2012-06-01 09:29:17] [8c387ce24444dc2c0e7d576cfc7658e0] - RMP [Histogram, QQplot and Density] [plot qq] [2012-08-30 09:47:14] [74be16979710d4c4e7c6647856088456] - R PD [Histogram, QQplot and Density] [] [2012-08-30 10:00:56] [e5cd290daa2365fa501ea04dbf3bfa28] - R P [Histogram, QQplot and Density] [Ian test] [2013-05-07 10:35:00] [98fd0e87c3eb04e0cc2efde01dbafab6] - RMPD [Wilcoxon-Mann-Whitney Test] [Heartbeat vs. NoH...] [2013-05-09 09:20:45] [873a05c15549159fecd09e71c73360f4] - R P [Histogram, QQplot and Density] [] [2013-05-09 09:23:58] [74be16979710d4c4e7c6647856088456] [Truncated] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
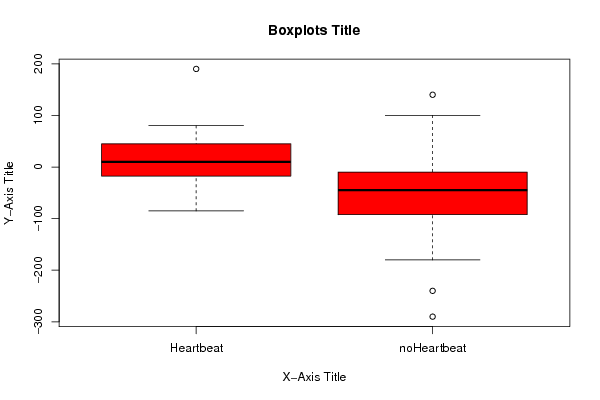
190 140 80 100 80 100 75 70 50 25 40 20 30 10 20 0 20 -10 10 -10 10 -25 10 -25 0 -25 0 -30 -10 -30 -25 -30 -30 -45 -45 -45 -60 -45 -85 -50 NA -50 NA -50 NA -60 NA -75 NA -75 NA -85 NA -85 NA -100 NA -110 NA -130 NA -130 NA -155 NA -155 NA -180 NA -240 NA -290 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = two.sided ; par2 = 1 ; par3 = 2 ; par4 = T-Test ; par5 = unpaired ; par6 = 0.0 ; par7 = 0.95 ; par8 = TRUE ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = two.sided ; par2 = 1 ; par3 = 2 ; par4 = T-Test ; par5 = unpaired ; par6 = 0.0 ; par7 = 0.95 ; par8 = TRUE ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
par2 <- as.numeric(par2) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||