Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_chi_squared_tests.wasp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Title produced by software | Chi-Squared and McNemar Tests | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Wed, 10 Nov 2010 15:53:42 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2010/Nov/10/t1289404375dea9nblds22axqt.htm/, Retrieved Mon, 29 Apr 2024 12:56:36 +0000 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=93093, Retrieved Mon, 29 Apr 2024 12:56:36 +0000 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 240 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| - [ECEL 2008] [ECEL2008 hypothes...] [2008-06-14 21:17:24] [74be16979710d4c4e7c6647856088456] - RMPD [Chi-Squared and McNemar Tests] [Chi squared 1] [2010-11-05 22:27:37] [97ad38b1c3b35a5feca8b85f7bc7b3ff] F R PD [Chi-Squared and McNemar Tests] [Connected versus ...] [2010-11-10 15:53:42] [dc77c696707133dea0955379c56a2acd] [Current] F [Chi-Squared and McNemar Tests] [Happiness versus ...] [2010-11-10 16:00:07] [95e8426e0df851c9330605aa1e892ab5] - R P [Chi-Squared and McNemar Tests] [WS 6 Connected vs...] [2010-11-12 14:58:54] [7b479c2bada71feddb7d988499871dfc] - R P [Chi-Squared and McNemar Tests] [WS 6 Connected vs...] [2010-11-12 15:01:40] [7b479c2bada71feddb7d988499871dfc] - R P [Chi-Squared and McNemar Tests] [WS 6 Separated vs...] [2010-11-12 15:03:56] [7b479c2bada71feddb7d988499871dfc] - R P [Chi-Squared and McNemar Tests] [WS 6 Separated vs...] [2010-11-12 15:05:21] [7b479c2bada71feddb7d988499871dfc] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'LO' 'LO' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'LO' 'LO' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'LO' 'LO' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'LO' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'LO' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'LO' 'LO' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'LO' 'LO' 'LO' 'LO' 'LO' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'LO' 'LO' 'LO' 'LO' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'LO' 'HI' 'LO' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'LO' 'LO' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'HI' 'HI' 'LO' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' 'HI' 'LO' 'HI' 'LO' 'LO' 'LO' 'LO' 'HI' 'LO' 'HI' 'HI' 'HI' 'LO' 'HI' | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
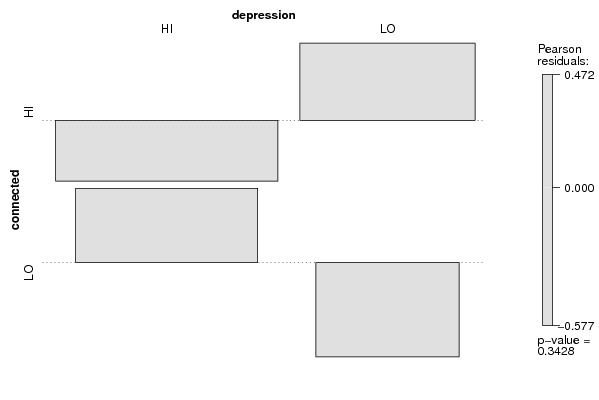
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = 3 ; par3 = Pearson Chi-Squared ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| par1 = 1 ; par2 = 4 ; par3 = Pearson Chi-Squared ; | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
library(vcd) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||