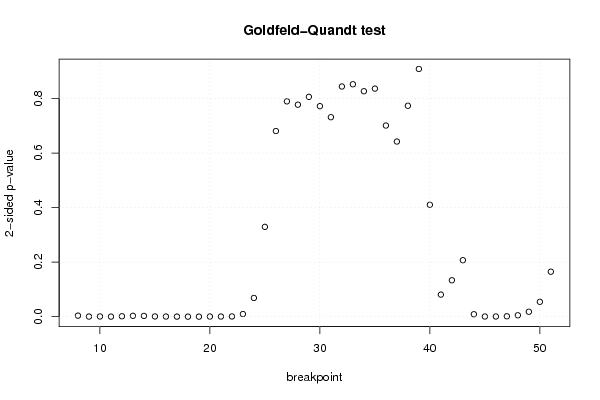
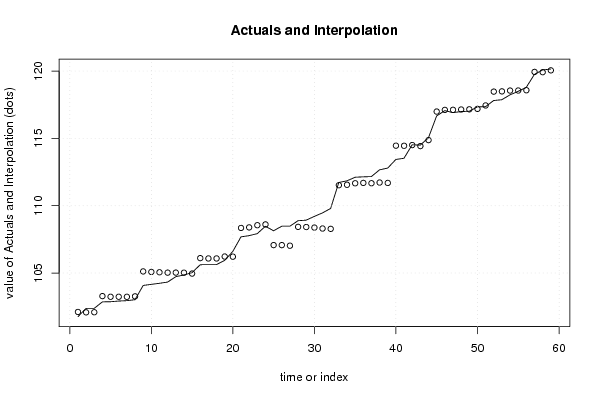
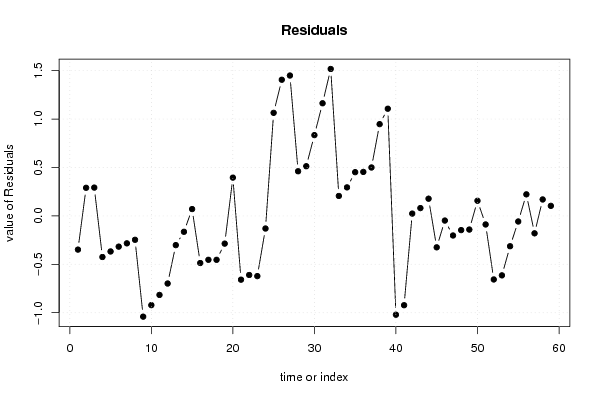
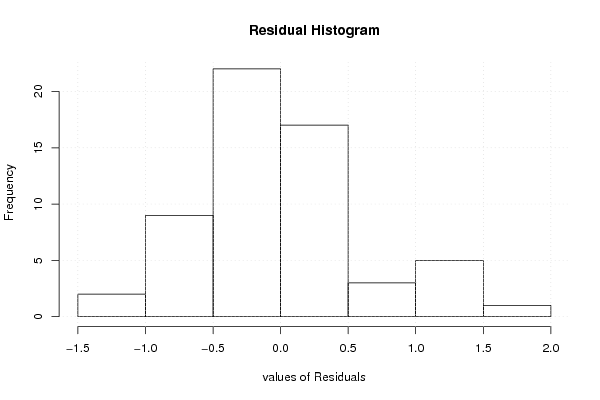
101,82 107,34 93,63 99,85 101,76
101,68 107,34 93,63 99,91 102,37
101,68 107,34 93,63 99,87 102,38
102,45 107,34 96,13 99,86 102,86
102,45 107,34 96,13 100,10 102,87
102,45 107,34 96,13 100,10 102,92
102,45 107,34 96,13 100,12 102,95
102,45 107,34 96,13 99,95 103,02
102,45 112,60 96,13 99,94 104,08
102,52 112,60 96,13 100,18 104,16
102,52 112,60 96,13 100,31 104,24
102,85 112,60 96,13 100,65 104,33
102,85 112,61 96,13 100,65 104,73
102,85 112,61 96,13 100,69 104,86
103,25 112,61 96,13 101,26 105,03
103,25 112,61 98,73 101,26 105,62
103,25 112,61 98,73 101,38 105,63
103,25 112,61 98,73 101,38 105,63
104,45 112,61 98,73 101,38 105,94
104,45 112,61 98,73 101,44 106,61
104,45 118,65 98,73 101,40 107,69
104,80 118,65 98,73 101,40 107,78
104,80 118,65 98,73 100,58 107,93
105,29 118,65 98,73 100,58 108,48
105,29 114,29 98,73 100,58 108,14
105,29 114,29 98,73 100,59 108,48
105,29 114,29 98,73 100,81 108,48
106,04 114,29 101,67 100,75 108,89
105,94 114,29 101,67 100,75 108,93
105,94 114,29 101,67 100,96 109,21
105,94 114,29 101,67 101,31 109,47
106,28 114,29 101,67 101,64 109,80
106,48 123,33 101,67 101,46 111,73
107,19 123,33 101,67 101,73 111,85
108,14 123,33 101,67 101,73 112,12
108,22 123,33 101,67 101,64 112,15
108,22 123,33 101,67 101,77 112,17
108,61 123,33 101,67 101,74 112,67
108,61 123,33 101,67 101,89 112,80
108,61 123,33 107,94 101,89 113,44
108,61 123,33 107,94 101,93 113,53
109,06 123,33 107,94 101,93 114,53
109,06 123,33 107,94 102,32 114,51
112,93 123,33 107,94 102,41 115,05
115,84 129,03 107,94 103,58 116,67
118,57 128,76 107,94 104,12 117,07
118,57 128,76 107,94 104,10 116,92
118,86 128,76 107,94 104,15 117,00
118,98 128,76 107,94 104,15 117,02
119,27 128,76 107,94 104,16 117,35
119,39 128,76 107,94 102,94 117,36
119,49 128,76 110,30 103,07 117,82
119,59 128,76 110,30 103,04 117,88
120,12 128,76 110,30 103,06 118,24
120,14 128,76 110,30 103,05 118,50
120,14 128,76 110,30 102,95 118,80
120,14 132,63 110,30 102,95 119,76
120,14 132,63 110,30 103,05 120,09
120,62 132,63 110,30 102,65 120,16 |