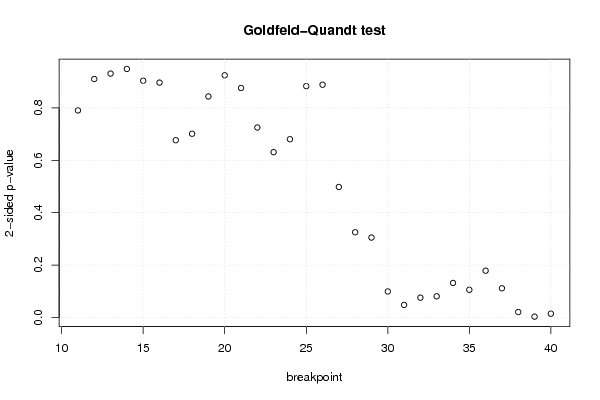
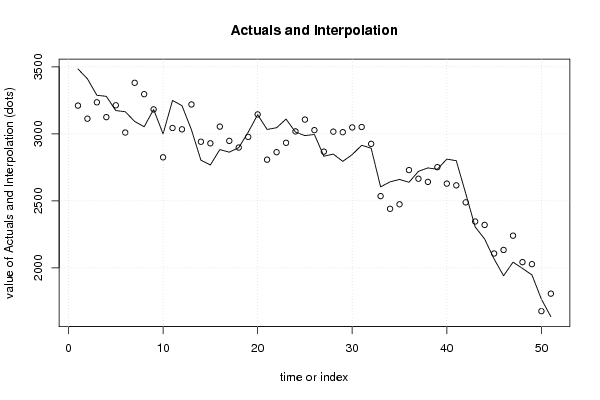
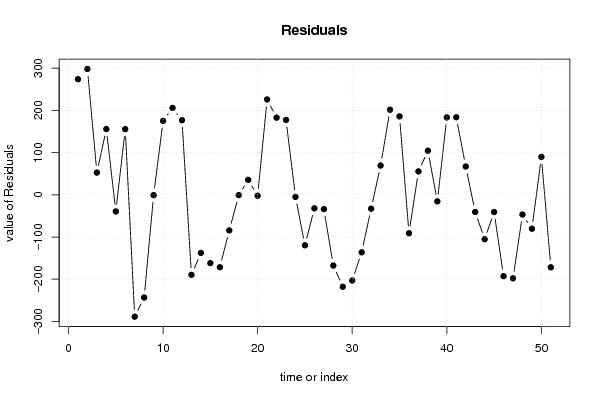
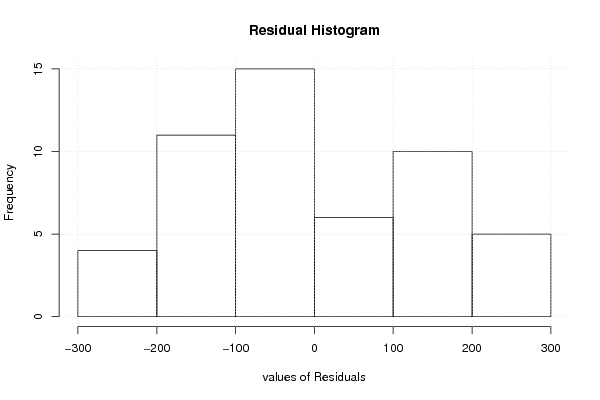
3484,74 13830,14 9349,44 7977 -5,6 6 1 2,77
3411,13 14153,22 9327,78 8241 -6,2 3 1 2,76
3288,18 15418,03 9753,63 8444 -7,1 2 1,2 2,76
3280,37 16666,97 10443,5 8490 -1,4 2 1,2 2,46
3173,95 16505,21 10853,87 8388 -0,1 2 0,8 2,46
3165,26 17135,96 10704,02 8099 -0,9 -8 0,7 2,47
3092,71 18033,25 11052,23 7984 0 0 0,7 2,71
3053,05 17671 10935,47 7786 0,1 -2 0,9 2,8
3181,96 17544,22 10714,03 8086 2,6 3 1,2 2,89
2999,93 17677,9 10394,48 9315 6 5 1,3 3,36
3249,57 18470,97 10817,9 9113 6,4 8 1,5 3,31
3210,52 18409,96 11251,2 9023 8,6 8 1,9 3,5
3030,29 18941,6 11281,26 9026 6,4 9 1,8 3,51
2803,47 19685,53 10539,68 9787 7,7 11 1,9 3,71
2767,63 19834,71 10483,39 9536 9,2 13 2,2 3,71
2882,6 19598,93 10947,43 9490 8,6 12 2,1 3,71
2863,36 17039,97 10580,27 9736 7,4 13 2,2 4,21
2897,06 16969,28 10582,92 9694 8,6 15 2,7 4,21
3012,61 16973,38 10654,41 9647 6,2 13 2,8 4,21
3142,95 16329,89 11014,51 9753 6 16 2,9 4,5
3032,93 16153,34 10967,87 10070 6,6 10 3,4 4,51
3045,78 15311,7 10433,56 10137 5,1 14 3 4,51
3110,52 14760,87 10665,78 9984 4,7 14 3,1 4,51
3013,24 14452,93 10666,71 9732 5 15 2,5 4,32
2987,1 13720,95 10682,74 9103 3,6 13 2,2 4,02
2995,55 13266,27 10777,22 9155 1,9 8 2,3 4,02
2833,18 12708,47 10052,6 9308 -0,1 7 2,1 3,85
2848,96 13411,84 10213,97 9394 -5,7 3 2,8 3,84
2794,83 13975,55 10546,82 9948 -5,6 3 3,1 4,02
2845,26 12974,89 10767,2 10177 -6,4 4 2,9 3,82
2915,02 12151,11 10444,5 10002 -7,7 4 2,6 3,75
2892,63 11576,21 10314,68 9728 -8 0 2,7 3,74
2604,42 9996,83 9042,56 10002 -11,9 -4 2,3 3,14
2641,65 10438,9 9220,75 10063 -15,4 -14 2,3 2,91
2659,81 10511,22 9721,84 10018 -15,5 -18 2,1 2,84
2638,53 10496,2 9978,53 9960 -13,4 -8 2,2 2,85
2720,25 10300,79 9923,81 10236 -10,9 -1 2,9 2,85
2745,88 9981,65 9892,56 10893 -10,8 1 2,6 3,08
2735,7 11448,79 10500,98 10756 -7,3 2 2,7 3,3
2811,7 11384,49 10179,35 10940 -6,5 0 1,8 3,29
2799,43 11717,46 10080,48 10997 -5,1 1 1,3 3,26
2555,28 10965,88 9492,44 10827 -5,3 0 0,9 3,26
2304,98 10352,27 8616,49 10166 -6,8 -1 1,3 3,11
2214,95 9751,2 8685,4 10186 -8,4 -3 1,3 2,84
2065,81 9354,01 8160,67 10457 -8,4 -3 1,3 2,71
1940,49 8792,5 8048,1 10368 -9,7 -3 1,3 2,69
2042 8721,14 8641,21 10244 -8,8 -4 1,1 2,65
1995,37 8692,94 8526,63 10511 -9,6 -8 1,4 2,57
1946,81 8570,73 8474,21 10812 -11,5 -9 1,2 2,32
1765,9 8538,47 7916,13 10738 -11 -13 1,7 2,12
1635,25 8169,75 7977,64 10171 -14,9 -18 1,8 2,05 |