Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_boxcoxnorm.wasp | ||||||||||||||||||||||||||||||||||||
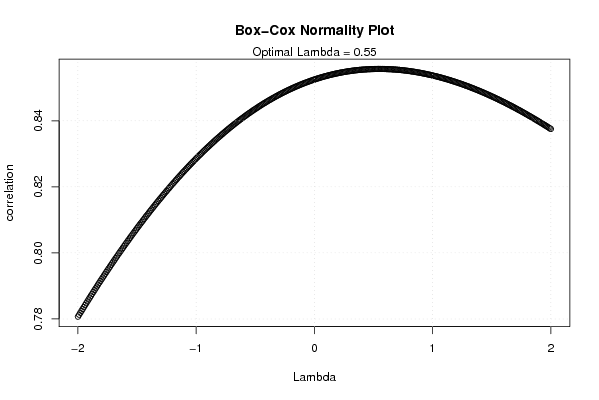
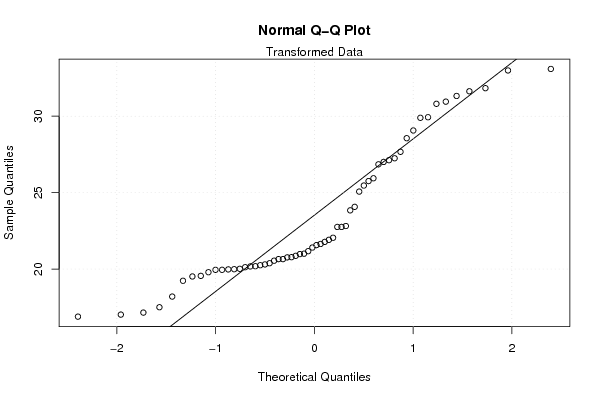
| Title produced by software | Box-Cox Normality Plot | ||||||||||||||||||||||||||||||||||||
| Date of computation | Thu, 13 Nov 2008 04:16:25 -0700 | ||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2008/Nov/13/t12265750501l95hkmh3e7nyjw.htm/, Retrieved Sat, 05 Jul 2025 18:20:28 +0000 | ||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=24572, Retrieved Sat, 05 Jul 2025 18:20:28 +0000 | |||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||
| User-defined keywords | Eigen Tijdreeks H8 Q4 | ||||||||||||||||||||||||||||||||||||
| Estimated Impact | 276 | ||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||
| F [Box-Cox Normality Plot] [loïqueverhasselt] [2008-11-13 11:16:25] [6440ec5a21e5d35520cb2ae6b4b70e45] [Current] F D [Box-Cox Normality Plot] [] [2008-11-13 16:08:26] [0dbd86b1fa2b13613c640b8c253471b2] F RMPD [Testing Mean with known Variance - Critical Value] [] [2008-11-13 16:16:13] [0dbd86b1fa2b13613c640b8c253471b2] F RMPD [Testing Mean with known Variance - p-value] [] [2008-11-13 16:19:46] [0dbd86b1fa2b13613c640b8c253471b2] F RMPD [Testing Mean with known Variance - Type II Error] [] [2008-11-13 16:25:35] [0dbd86b1fa2b13613c640b8c253471b2] F RMPD [Testing Mean with known Variance - Sample Size] [] [2008-11-13 16:28:33] [0dbd86b1fa2b13613c640b8c253471b2] F RMPD [Testing Population Mean with known Variance - Confidence Interval] [] [2008-11-13 16:30:58] [0dbd86b1fa2b13613c640b8c253471b2] F RMPD [Testing Population Mean with known Variance - Confidence Interval] [] [2008-11-13 16:33:00] [0dbd86b1fa2b13613c640b8c253471b2] | |||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||
93 98,4 92,6 94,6 99,5 97,6 91,3 93,6 93,1 78,4 70,2 69,3 71,1 73,5 85,9 91,5 91,8 88,3 91,3 94 99,3 96,7 88 96,7 106,8 114,3 105,7 90,1 91,6 97,7 100,8 104,6 95,9 102,7 104 107,9 113,8 113,8 123,1 125,1 137,6 134 140,3 152,1 150,6 167,3 153,2 142 154,4 158,5 180,9 181,3 172,4 192 199,3 215,4 214,3 201,5 190,5 196 | |||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||
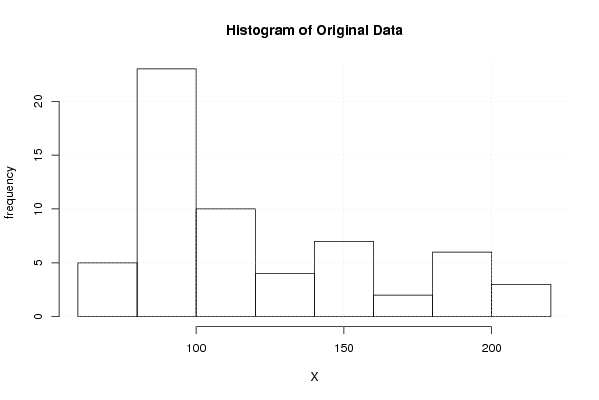
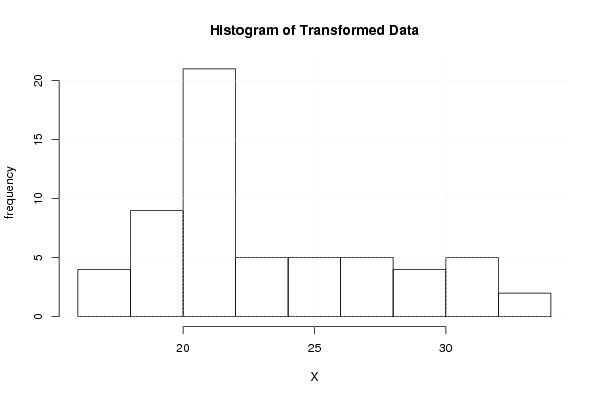
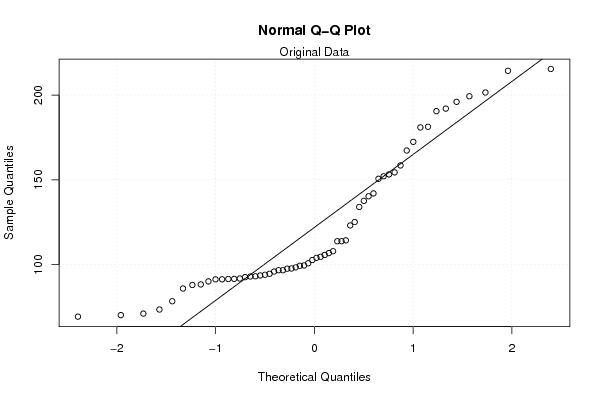
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||
n <- length(x) | |||||||||||||||||||||||||||||||||||||