Free Statistics
of Irreproducible Research!
Description of Statistical Computation | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Author's title | |||||||||||||||||||||||||||||||||||||||||||||
| Author | *The author of this computation has been verified* | ||||||||||||||||||||||||||||||||||||||||||||
| R Software Module | rwasp_bidensity.wasp | ||||||||||||||||||||||||||||||||||||||||||||
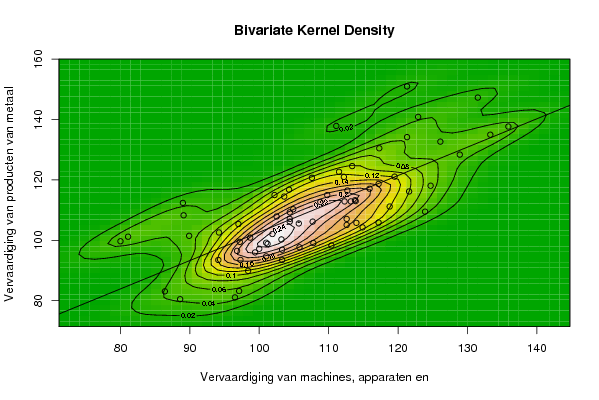
| Title produced by software | Bivariate Kernel Density Estimation | ||||||||||||||||||||||||||||||||||||||||||||
| Date of computation | Mon, 10 Nov 2008 02:23:45 -0700 | ||||||||||||||||||||||||||||||||||||||||||||
| Cite this page as follows | Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?v=date/2008/Nov/10/t1226309091q4yvbfmqmpv20i6.htm/, Retrieved Wed, 09 Jul 2025 06:39:04 +0000 | ||||||||||||||||||||||||||||||||||||||||||||
| Statistical Computations at FreeStatistics.org, Office for Research Development and Education, URL https://freestatistics.org/blog/index.php?pk=22892, Retrieved Wed, 09 Jul 2025 06:39:04 +0000 | |||||||||||||||||||||||||||||||||||||||||||||
| QR Codes: | |||||||||||||||||||||||||||||||||||||||||||||
|
| |||||||||||||||||||||||||||||||||||||||||||||
| Original text written by user: | |||||||||||||||||||||||||||||||||||||||||||||
| IsPrivate? | No (this computation is public) | ||||||||||||||||||||||||||||||||||||||||||||
| User-defined keywords | |||||||||||||||||||||||||||||||||||||||||||||
| Estimated Impact | 276 | ||||||||||||||||||||||||||||||||||||||||||||
Tree of Dependent Computations | |||||||||||||||||||||||||||||||||||||||||||||
| Family? (F = Feedback message, R = changed R code, M = changed R Module, P = changed Parameters, D = changed Data) | |||||||||||||||||||||||||||||||||||||||||||||
| F [Univariate Data Series] [VMAW] [2008-10-13 16:54:40] [cbd3d88cd5aad6543e769146e7e26b0c] F RMPD [Bivariate Kernel Density Estimation] [Opdracht 4 Q1] [2008-11-10 09:23:45] [2ae704d6b0222e84f58032588d68322b] [Current] - RMPD [Partial Correlation] [] [2008-11-11 16:13:50] [3f66c6f083b1153972739491b89fa2dd] - RMPD [Hierarchical Clustering] [sarah Q2] [2008-11-11 16:29:51] [3f66c6f083b1153972739491b89fa2dd] - RMPD [Hierarchical Clustering] [sarah Q2] [2008-11-11 16:37:31] [3f66c6f083b1153972739491b89fa2dd] F RMPD [Testing Mean with known Variance - Critical Value] [] [2008-11-11 17:01:20] [3f66c6f083b1153972739491b89fa2dd] | |||||||||||||||||||||||||||||||||||||||||||||
| Feedback Forum | |||||||||||||||||||||||||||||||||||||||||||||
Post a new message | |||||||||||||||||||||||||||||||||||||||||||||
Dataset | |||||||||||||||||||||||||||||||||||||||||||||
| Dataseries X: | |||||||||||||||||||||||||||||||||||||||||||||
97.3 101 113.2 101 105.7 113.9 86.4 96.5 103.3 114.9 105.8 94.2 98.4 99.4 108.8 112.6 104.4 112.2 81.1 97.1 112.6 113.8 107.8 103.2 103.3 101.2 107.7 110.4 101.9 115.9 89.9 88.6 117.2 123.9 100 103.6 94.1 98.7 119.5 112.7 104.4 124.7 89.1 97 121.6 118.8 114 111.5 97.2 102.5 113.4 109.8 104.9 126.1 80 96.8 117.2 112.3 117.3 111.1 102.2 104.3 122.9 107.6 121.3 131.5 89 104.4 128.9 135.9 133.3 121.3 | |||||||||||||||||||||||||||||||||||||||||||||
| Dataseries Y: | |||||||||||||||||||||||||||||||||||||||||||||
93.5 94.7 112.9 99.2 105.6 113 83.1 81.1 96.9 104.3 97.7 102.6 89.9 96 112.7 107.1 106.2 121 101.2 83.2 105.1 113.3 99.1 100.3 93.5 98.8 106.2 98.3 102.1 117.1 101.5 80.5 105.9 109.5 97.2 114.5 93.5 100.9 121.1 116.5 109.3 118.1 108.3 105.4 116.2 111.2 105.8 122.7 99.5 107.9 124.6 115 110.3 132.7 99.7 96.5 118.7 112.9 130.5 137.9 115 116.8 140.9 120.7 134.2 147.3 112.4 107.1 128.4 137.7 135 151 | |||||||||||||||||||||||||||||||||||||||||||||
Tables (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||
Figures (Output of Computation) | |||||||||||||||||||||||||||||||||||||||||||||
Input Parameters & R Code | |||||||||||||||||||||||||||||||||||||||||||||
| Parameters (Session): | |||||||||||||||||||||||||||||||||||||||||||||
| par1 = 50 ; par2 = 50 ; par3 = 0 ; par4 = 0 ; par5 = 0 ; par6 = Y ; par7 = Y ; | |||||||||||||||||||||||||||||||||||||||||||||
| Parameters (R input): | |||||||||||||||||||||||||||||||||||||||||||||
| par1 = 50 ; par2 = 50 ; par3 = 0 ; par4 = 0 ; par5 = 0 ; par6 = Y ; par7 = Y ; | |||||||||||||||||||||||||||||||||||||||||||||
| R code (references can be found in the software module): | |||||||||||||||||||||||||||||||||||||||||||||
par1 <- as(par1,'numeric') | |||||||||||||||||||||||||||||||||||||||||||||